2MP9
 
 | | Solution structure of an potent antifungal peptide Cm-p5 derived from C. muricatus | | Descriptor: | Antifungal peptide | | Authors: | Sun, Z.J, Heffron, G, Mcbeth, C, Wagner, G, Otero-Gonzales, A.J, Starnbach, M.N. | | Deposit date: | 2014-05-14 | | Release date: | 2015-08-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cm-p5: an antifungal hydrophilic peptide derived from the coastal mollusk Cenchritis muricatus (Gastropoda: Littorinidae).
Faseb J., 29, 2015
|
|
1ZGW
 
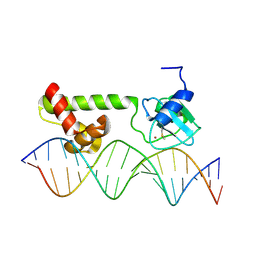 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1KOY
 
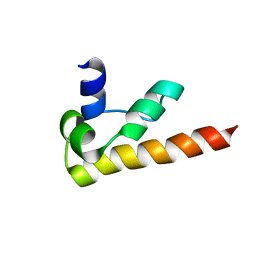 | | NMR structure of DFF-C domain | | Descriptor: | DNA fragmentation factor alpha subunit | | Authors: | Fukushima, K, Kikuchi, J, Koshiba, S, Kigawa, T, Kuroda, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-25 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation.
J.Mol.Biol., 321, 2002
|
|
2KC3
 
 | |
2K21
 
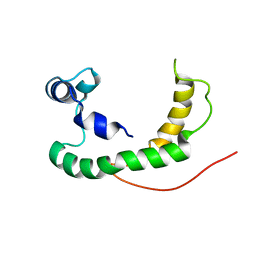 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2LQC
 
 | |
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
2LMT
 
 | | NMR structure of Androcam | | Descriptor: | CALCIUM ION, Calmodulin-related protein 97A | | Authors: | Joshi, M.K, Moran, S.T, Beckingham, K.M, Mackenzie, K.R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of androcam supports specialized interactions with myosin VI.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LIR
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in oxidized states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
2LW9
 
 | | NMR solution structure of Myo10 anti-CC | | Descriptor: | Unconventionnal myosin-X | | Authors: | Ye, F, Lu, Q, Zhang, M. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Antiparallel coiled-coil-mediated dimerization of myosin X
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MH3
 
 | | NMR structure of the basic helix-loop-helix region of the transcriptional repressor HES-1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Wienk, H, Popovic, M, Coglievina, M, Boelens, R, Pongor, S, Pintar, A. | | Deposit date: | 2013-11-15 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The basic helix-loop-helix region of the transcriptional repressor hairy and enhancer of split 1 is preorganized to bind DNA.
Proteins, 82, 2014
|
|
2MLI
 
 | |
2MWA
 
 | |
2MX4
 
 | | NMR structure of Phosphorylated 4E-BP2 | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 2 | | Authors: | Bah, A, Forman-Kay, J, Vernon, R, Siddiqui, Z, Krzeminski, M, Muhandiram, R, Zhao, C, Sonenberg, N, Kay, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-07 | | Last modified: | 2015-03-18 | | Method: | SOLUTION NMR | | Cite: | Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch.
Nature, 519, 2015
|
|
2N0C
 
 | | NMR structure of Neuromedin C in 10% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
1IC9
 
 | |
2N0B
 
 | | NMR structure of Neuromedin C in aqueous solution | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2MWF
 
 | |
2N0F
 
 | | NMR structure of Neuromedin C in 60% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0G
 
 | | NMR structure of Neuromedin C in 90% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2MWD
 
 | |
2MW9
 
 | |
2MWE
 
 | | NMR structure of FBP28 WW2 mutant Y438R, L453A DNDC | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1KCY
 
 | | NMR solution structure of apo calbindin D9k (F36G + P43M mutant) | | Descriptor: | calbindin D9k | | Authors: | Nelson, M.R, Thulin, E, Fagan, P.A, Forsen, S, Chazin, W.J. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The EF-hand domain: a globally cooperative structural unit.
Protein Sci., 11, 2002
|
|
1KMF
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
