7MFQ
 
 | | crystal structure of the L136 aminotransferase from acanthamoeba polyphaga mimivirus in complex with the TDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
4ZFZ
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with myristoylated 5-mer lipopeptide derived from SIV Nef protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-mer lipopeptide from Protein Nef, ... | | Authors: | Morita, D, Sugita, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of the N-myristoylated lipopeptide-bound MHC class I complex
Nat Commun, 7, 2016
|
|
8AWK
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with D-carbaxylosyl chloride | | Descriptor: | (2~{S},3~{S},4~{R})-cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8GLF
 
 | | Crystal Structure of Human CD1b in Complex with Sphingomyelin C34:2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Shahine, A. | | Deposit date: | 2023-03-22 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Cell, 186, 2023
|
|
9H65
 
 | | Steroidal Selective Modulators of FXR with Therapeutic Potential | | Descriptor: | 1-[(3R)-3-[(3R,4R,5S,6R,7R,8S,9S,10R,13R,14S,17R)-6-ethyl-4-fluoranyl-10,13-dimethyl-3,7-bis(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]butyl]-3-(phenylsulfonyl)urea, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Kydd-Sinclair, D, Watson, K.A. | | Deposit date: | 2024-10-23 | | Release date: | 2025-09-10 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Novel Bile Acid-Based Modulators of FXR.
J.Mol.Biol., 437, 2025
|
|
5TQA
 
 | |
3IAE
 
 | | Structure of benzaldehyde lyase A28S mutant with benzoylphosphonate | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzaldehyde lyase, CALCIUM ION | | Authors: | Brandt, G.S, Petsko, G.A, Ringe, D, McLeish, M.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site engineering of benzaldehyde lyase shows that a point mutation can confer both new reactivity and susceptibility to mechanism-based inhibition.
J.Am.Chem.Soc., 132, 2010
|
|
3N0W
 
 | |
4QYX
 
 | | Crystal structure of YDR533Cp | | Descriptor: | Probable chaperone protein HSP31 | | Authors: | Wilson, M.A, Amour, S.T, Collins, J.L, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8GLG
 
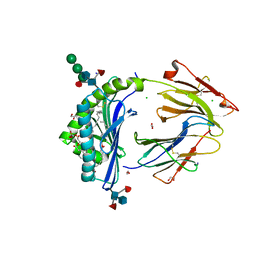 | | Crystal Structure of Human CD1b in Complex with Phosphatidylethanolamine C34:1 | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shahine, A. | | Deposit date: | 2023-03-22 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Cell, 186, 2023
|
|
1KY1
 
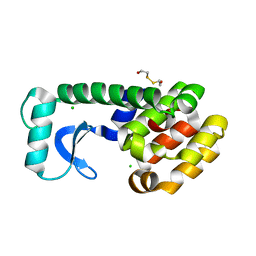 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
7ZFS
 
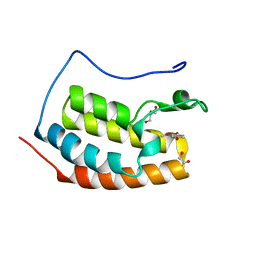 | | BRD4 in complex with FragLite32 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1,2-dimethoxy-benzene, Bromodomain-containing protein 4, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7HHS
 
 | |
7HCA
 
 | |
7D66
 
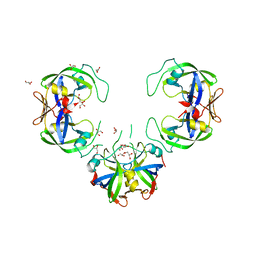 | |
6SKR
 
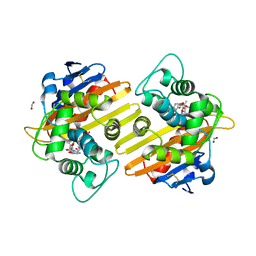 | | OXA-10_ETP. Structural insight to the enhanced carbapenem efficiency of OXA-655 compared to OXA-10. | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, FORMIC ACID, ... | | Authors: | Leiros, H.-K.S. | | Deposit date: | 2019-08-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the enhanced carbapenemase efficiency of OXA-655 compared to OXA-10.
Febs Open Bio, 10, 2020
|
|
9ATZ
 
 | | HIV 16055.v8.3 SOSIP Env in Complex with V2 Epitope and Anti-Immune Complex pAbs from Rabbit 2464 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rabbit Anti-Immune Complex Antibody - Predicted Heavy Chain, ... | | Authors: | Brown, S, Antansijevic, A, Ward, A.B. | | Deposit date: | 2024-02-27 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Anti-immune complex antibodies are elicited during repeated immunization with HIV Env immunogens.
Sci Immunol, 10, 2025
|
|
3CJ5
 
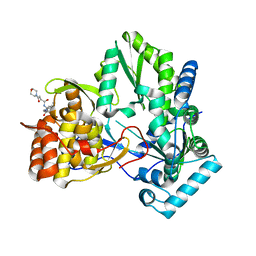 | |
6HTD
 
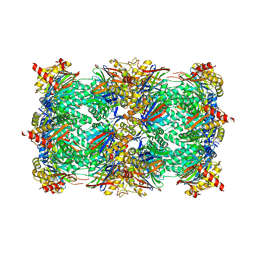 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 4 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-03 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
1L67
 
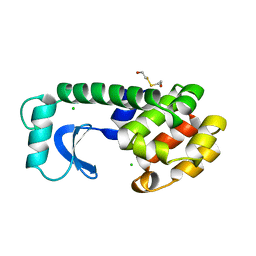 | |
6HV3
 
 | | Yeast 20S proteasome with human beta2i (1-53) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
9E6R
 
 | |
9EJG
 
 | | Peptide-independent T cell receptor recognition of HLA-DQ2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, FORMIC ACID, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A naturally selected alpha beta T cell receptor binds HLA-DQ2 molecules without co-contacting the presented peptide.
Nat Commun, 16, 2025
|
|
2YF3
 
 | | Crystal structure of DR2231, the MazG-like protein from Deinococcus radiodurans, complex with manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE, ... | | Authors: | Goncalves, A.M.D, deSanctis, D, McSweeney, S.M. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
1L16
 
 | |
