4CSR
 
 | | High resolution crystal structure of the histone fold dimer (NF-YB)-(NF-YC) | | Descriptor: | GLYCEROL, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT BETA, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT GAMMA | | Authors: | Gnesutta, N, Cocolo, S, Mantovani, R, Bolognesi, M, Nardini, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Crystal Structure of the Histone Fold Dimer (NF-Yb)-(NF-Yc)
To be Published
|
|
7VDV
 
 | | The overall structure of human chromatin remodeling PBAF-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2,AT-rich interactive domain-containing protein 2, Actin, ... | | Authors: | Chen, Z.C, Chen, K.J, Yuan, J.J. | | Deposit date: | 2021-09-07 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human chromatin-remodelling PBAF complex bound to a nucleosome.
Nature, 605, 2022
|
|
4CO8
 
 | | Structure of the DNA binding ETS domain of human ETV4 | | Descriptor: | 1,2-ETHANEDIOL, ETS TRANSLOCATION VARIANT 4 | | Authors: | Newman, J.A, Cooper, C.D.O, Shrestha, L, Burgess-Brown, N, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2014-01-27 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4D1X
 
 | | CDK2 in complex with Luciferin | | Descriptor: | (4S)-2-(6-hydroxy-1,3-benzothiazol-2-yl)-4,5-dihydro-1,3-thiazole-4-carboxylic acid, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Rothweiler, U, Engh, R.A. | | Deposit date: | 2014-05-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Luciferin and Derivatives as a Dyrk Selective Scaffold for the Design of Protein Kinase Inhibitors.
Eur.J.Med.Chem., 94, 2015
|
|
4CZZ
 
 | | Histone demethylase LSD1(KDM1A)-CoREST3 Complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 3 | | Authors: | Barrios, A.P, Gomez, A.V, Saez, J.E, Ciossani, G, Toffolo, E, Battaglioli, E, Mattevi, A, Andres, M.E. | | Deposit date: | 2014-04-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Properties of Transcriptional Complexes Formed by the Corest Family.
Mol.Cell.Biol., 34, 2014
|
|
5N7G
 
 | | MAGI-1 complexed with a synthetic pRSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
8VG2
 
 | |
8GQ6
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H35
 
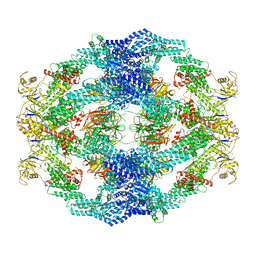 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.41 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H33
 
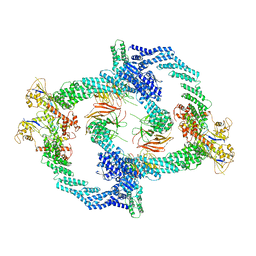 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.86 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.99 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5N7F
 
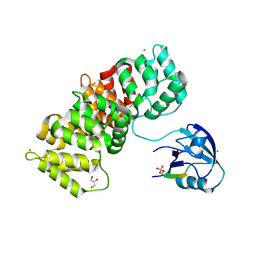 | | MAGI-1 complexed with a pRSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
5N7D
 
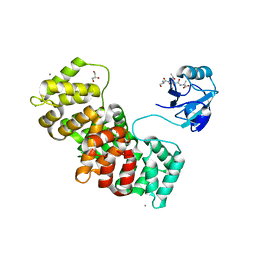 | | MAGI-1 complexed with a RSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
2V5W
 
 | | Crystal structure of HDAC8-substrate complex | | Descriptor: | GLYCYL-GLYCYL-GLYCINE, HISTONE DEACETYLASE 8, PEPTIDIC SUBSTRATE, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
3D87
 
 | | Crystal structure of Interleukin-23 | | Descriptor: | Interleukin-12 subunit p40, Interleukin-23 subunit p19, PHOSPHATE ION, ... | | Authors: | Beyer, B.M, Ingram, R, Ramanathan, L, Reichert, P, Le, H, Madison, V. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody
J.Mol.Biol., 382, 2008
|
|
2UZE
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(Z)-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]FURAN-2-YL}BENZOIC ACID, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2V7D
 
 | |
5D2D
 
 | | Crystal structure of human 14-3-3 zeta in complex with CFTR R-domain peptide pS753-pS768 | | Descriptor: | 14-3-3 protein zeta/delta, CHLORIDE ION, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Stevers, L.M, Leysen, S.F.R, Ottmann, C. | | Deposit date: | 2015-08-05 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3D85
 
 | | Crystal structure of IL-23 in complex with neutralizing FAB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FAB of antibody 7G10, heavy chain, ... | | Authors: | Beyer, B.M, Ingram, R, Ramanathan, L, Reichert, P, Le, H, Madison, V. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody
J.Mol.Biol., 382, 2008
|
|
5D3F
 
 | |
2V5X
 
 | | Crystal structure of HDAC8-inhibitor complex | | Descriptor: | (2R)-N~8~-HYDROXY-2-{[(5-METHOXY-2-METHYL-1H-INDOL-3-YL)ACETYL]AMINO}-N~1~-[2-(2-PHENYL-1H-INDOL-3-YL)ETHYL]OCTANEDIAMIDE, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C, Gallinari, P, Jones, P, Mattu, M, Carfi, A, Defrancesco, R, Steinkuhler, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
