7YX5
 
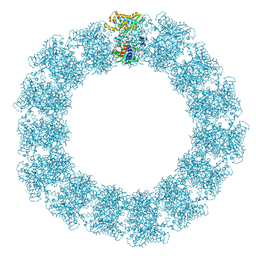 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
5ABJ
 
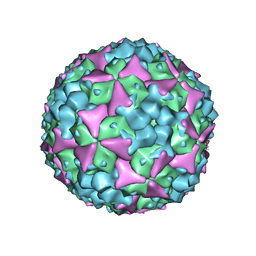 | | Structure of Coxsackievirus A16 in complex with GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
9FD8
 
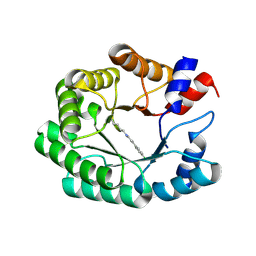 | | Re-engineered peroxygenase variant of 2-deoxy-D-ribose-5-phosphate aldolase, Schiff-base complex with 4-chloro-cinnamaldehyde | | Descriptor: | (2E)-3-(4-chlorophenyl)prop-2-enal, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Zhou, H, Frietema, H.O.T, Poelarends, G.J. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Engineering 2-Deoxy-D-ribose-5-phosphate Aldolase for anti- and syn-Selective Epoxidations of alpha , beta-Unsaturated Aldehydes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6I23
 
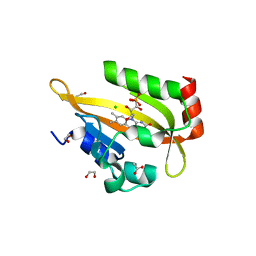 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
4U5D
 
 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5757 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
6P9K
 
 | |
9FD9
 
 | | Re-engineered peroxygenase variant of 2-deoxy-D-ribose-5-phosphate aldolase, Schiff-base complex with 4-nitro-cinnamaldehyde | | Descriptor: | 4-nitro-cinnamaldehyde, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Zhou, H, Frietema, H.O.T, Poelarends, G.J. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Engineering 2-Deoxy-D-ribose-5-phosphate Aldolase for anti- and syn-Selective Epoxidations of alpha , beta-Unsaturated Aldehydes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7ZHA
 
 | |
6PD2
 
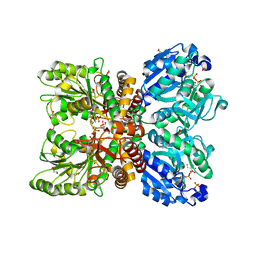 | |
3K1Z
 
 | | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Haloacid dehalogenase-like hydrolase domain-containing protein 3 | | Authors: | Ugochukwu, E, Guo, K, Yue, W.W, Pilka, E, Picaud, S, Muniz, J, Pike, A.C.W, Krojer, T, Gomes, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3)
To be Published
|
|
5FSM
 
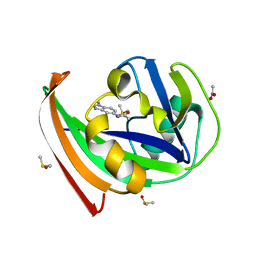 | | MTH1 substrate recognition: Complex with a methylbenzimidazolyl acetamide. | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
4U7Q
 
 | | Structure of wild-type HIV protease in complex with photosensitive inhibitor PDI-6 | | Descriptor: | N~2~-({[7-(diethylamino)-2-oxo-2H-chromen-4-yl]methoxy}carbonyl)-N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | Authors: | Pachl, P, Rezacova, P, Schimer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
4U7V
 
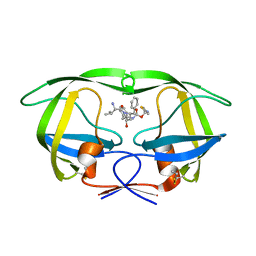 | | Structure of wild-type HIV protease in complex with degraded photosensitive inhibitor | | Descriptor: | BETA-MERCAPTOETHANOL, N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | Authors: | Pachl, P, Rezacova, P, Schimer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
1O13
 
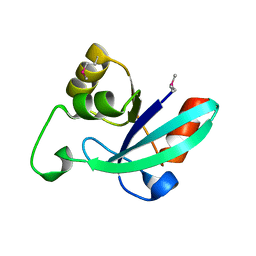 | |
5JXQ
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE (TGT) IN COMPLEX WITH 6-AMINO-2-{[4-(2-HYDROXYETHYL)PHENETHYL]AMINO}-1,7-DIHYDRO-8H-IMIDAZO[4,5-g]QUINAZOLIN-8-ONE | | Descriptor: | 6-amino-2-({2-[4-(2-hydroxyethyl)phenyl]ethyl}amino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Occupying a flat subpocket in a tRNA-modifying enzyme with ordered or disordered side chains: Favorable or unfavorable for binding?
Bioorg.Med.Chem., 24, 2016
|
|
5AE3
 
 | | Ether Lipid-Generating Enzyme AGPS in complex with antimycin A | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Piano, V, Benjamin, D.I, Valente, S, Nenci, S, Marrocco, B, Mai, A, Aliverti, A, Nomura, D.K, Mattevi, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Inhibitors for the Ether Lipid-Generating Enzyme Agps as Anti-Cancer Agents.
Acs Chem.Biol., 10, 2015
|
|
5FSN
 
 | | MTH1 substrate recognition: Complex with a aminomethylpyrimidinyl oxypropanol. | | Descriptor: | 3-(2-AMINO-6-METHYL-PYRIMIDIN-4-YL)OXYPROPAN-1-OL, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
5NKM
 
 | | SMG8-SMG9 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
5JTU
 
 | | Crystal structure of GPb in complex with 8b | | Descriptor: | (1S)-1,5-anhydro-1-[5-(naphthalen-2-yl)-1H-imidazol-2-yl]-D-glucitol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur.J.Med.Chem., 123, 2016
|
|
5AH0
 
 | | STRUCTURE OF LIPASE 1 FROM PELOSINUS FERMENTANS | | Descriptor: | DI(HYDROXYETHYL)ETHER, LIPASE, POTASSIUM ION, ... | | Authors: | Hromic, A, Gruber, K, Biundo, A, Ribitsch, D, Quartinello, F, Perz, V, Arrell, M.S, Kalman, F, Guebitz, G.M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a Poly(Butylene Adipate-Co-Terephthalate)-Hydrolyzing Lipase from Pelosinus Fermentans.
Appl.Microbiol.Biotechnol., 100, 2016
|
|
169L
 
 | |
9N7A
 
 | |
6C7Q
 
 | | BRD4 BD2 in complex with compound CE277 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-N-(1-methyl-1H-indazol-3-yl)-9H-pyrimido[4,5-b]indol-4-amine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
4TZ2
 
 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(5-phenyl-4H-1,2,4-triazol-3-yl)aniline, ATPase family AAA domain-containing protein 2, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
1A4V
 
 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Chandra, N, Acharya, K.R. | | Deposit date: | 1998-02-05 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for the presence of a secondary calcium binding site in human alpha-lactalbumin.
Biochemistry, 37, 1998
|
|
