6S63
 
 | | Dark-adapted structure of Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich at room temperature | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Axford, D, Bada Juarez, J.F, Vinals, J, Watts, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two states of a light-sensitive membrane protein captured at room temperature using thin-film sample mounts.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1MD8
 
 | | Monomeric structure of the active catalytic domain of complement protease C1r | | Descriptor: | C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
3HCJ
 
 | |
1MMI
 
 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7ZJJ
 
 | | CspZ (BbCRASP-2) from Borrelia burgdorferi strain B379 | | Descriptor: | CspZ, NITRATE ION | | Authors: | Brangulis, K, Marcinkiewicz, A, Hart, T.M, Dupuis, A.P, Zamba Campero, M, Nowak, T.A, Stout, J.L, Akopjana, I, Kazaks, A, Bogans, J, Ciota, A.T, Kraiczy, P, Kolokotronis, S.O, Lin, Y.-P. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7ZJM
 
 | | Crystal structure of a complex between CspZ from Borrelia burgdorferi strain B408 and human FH SCR domains 6-7 | | Descriptor: | Complement factor H, CspZ, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Brangulis, K, Marcinkiewicz, A, Hart, T.M, Dupuis, A.P, Zamba Campero, M, Nowak, T.A, Stout, J.L, Akopjana, I, Kazaks, A, Bogans, J, Ciota, A.T, Kraiczy, P, Kolokotronis, S.O, Lin, Y.-P. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7ZJK
 
 | | CspZ (BbCRASP-2) from Borrelia burgdorferi strain B408 | | Descriptor: | CspZ | | Authors: | Brangulis, K, Marcinkiewicz, A, Hart, T.M, Dupuis, A.P, Zamba Campero, M, Nowak, T.A, Stout, J.L, Akopjana, I, Kazaks, A, Bogans, J, Ciota, A.T, Kraiczy, P, Kolokotronis, S.O, Lin, Y.-P. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural evolution of an immune evasion determinant shapes pathogen host tropism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3LE2
 
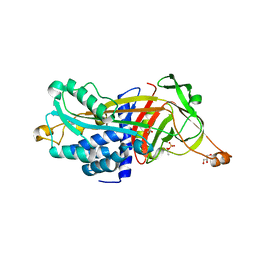 | | Structure of Arabidopsis AtSerpin1. Native Stressed Conformation | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Harrop, S.J, Joss, T.V, Cumi, P.M.G, Roberts, T.H. | | Deposit date: | 2010-01-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabidopsis AtSerpin1, crystal structure and in vivo interaction with its target protease RESPONSIVE TO DESICCATION-21 (RD21).
J.Biol.Chem., 285, 2010
|
|
7M90
 
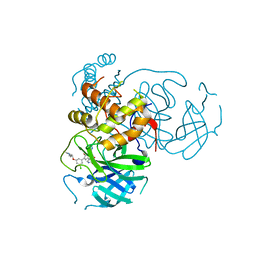 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 50 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8O
 
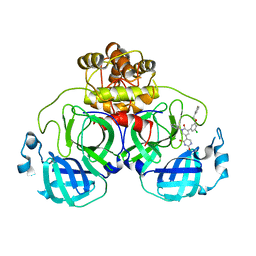 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 19 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M91
 
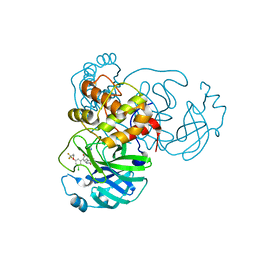 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 25 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8M
 
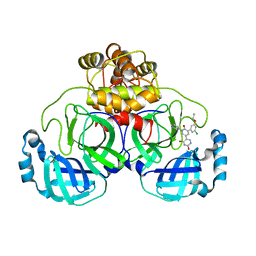 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 3C-like proteinase, 5-[3-(3-chloro-5-propoxyphenyl)-2-oxo-2H-[1,3'-bipyridin]-5-yl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Y
 
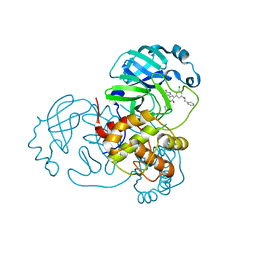 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 15 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8Z
 
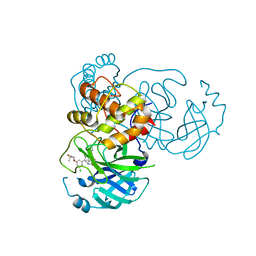 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 29 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3-hydroxy-3-methylbutoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8X
 
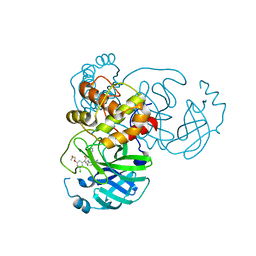 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 2-{3-[3-chloro-5-(2-methoxyethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8P
 
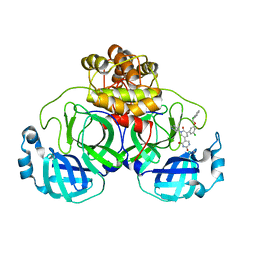 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 23 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7ZKU
 
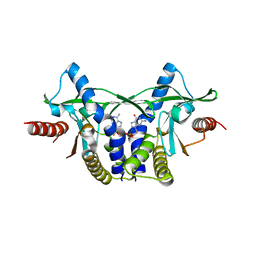 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'dGMP) | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R})-8-(6-aminopurin-9-yl)-9-fluoranyl-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]-2-azanyl-3~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-04-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'dGMP)
To Be Published
|
|
7M8N
 
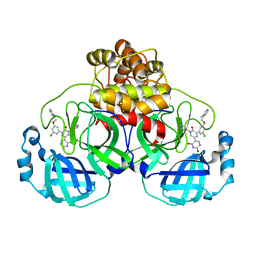 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 16 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(2-methylphenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7ZVK
 
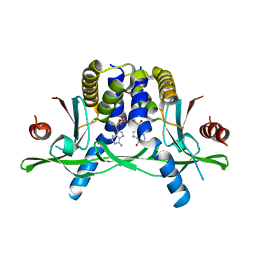 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-IMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-9-fluoranyl-3,12,18-tris(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-16 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-IMP)
To Be Published
|
|
5L2D
 
 | |
3QIM
 
 | | Histidine 416 of the periplamsic binding protein NikA is essential for nickel uptake in Escherichia coli | | Descriptor: | ACETATE ION, GLYCEROL, Nickel-binding periplasmic protein, ... | | Authors: | Cavazza, C, Martin, L, Laffly, E, Lebrette, H, Cherrier, M.V, Zeppieri, L, Richaud, P, Carriere, M, Fontecilla-Camps, J.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine 416 of the periplasmic binding protein NikA is essential for nickel uptake in Escherichia coli
Febs Lett., 585, 2011
|
|
3HSY
 
 | | High resolution structure of a dimeric GluR2 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, SULFATE ION, ... | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Greger, I.H. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
Embo J., 30, 2011
|
|
4KO3
 
 | | Low X-ray dose structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
3S97
 
 | | PTPRZ CNTN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Bouyain, S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2971 Å) | | Cite: | A complex between contactin-1 and the protein tyrosine phosphatase PTPRZ controls the development of oligodendrocyte precursor cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4KTM
 
 | | Crystal Structure of C143S Xanthomonas campestris OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION | | Authors: | Goblirsch, B.R. | | Deposit date: | 2013-05-20 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Substrate Trapping in Crystals of the Thiolase OleA Identifies Three Channels That Enable Long Chain Olefin Biosynthesis.
J.Biol.Chem., 291, 2016
|
|
