8EVD
 
 | |
8F6U
 
 | |
8F6V
 
 | |
5ZA2
 
 | | Fox-4 beta-lactamase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Nukaga, M, Hoshino, T, Papp-Wallace, K.M, Bonomo, R.A. | | Deposit date: | 2018-02-06 | | Release date: | 2018-03-07 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Probing the Mechanism of Inactivation of the FOX-4 Cephamycinase by Avibactam
Antimicrob. Agents Chemother., 62, 2018
|
|
8FQX
 
 | | Carbonic Anhydrase II in complex with the alkyl ureas 3g | | Descriptor: | 4-hydroxy-3-({[(pyridin-4-yl)methyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Combs, J.E, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FQZ
 
 | | Carbonic Anhydrase II in complex with the alkyl urea 3j | | Descriptor: | 4-hydroxy-3-{[(4-hydroxybutyl)carbamoyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FLP
 
 | | NMR Solution Structure of LvIC analogue | | Descriptor: | Alpha-conotoxin LvIC analogue | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Engineering of LvIC, an alpha 4/4-Conotoxin That Selectively Blocks Rat alpha 6/ alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 66, 2023
|
|
8FR1
 
 | | Carbonic Anhydrase IX in complex with the alkyl urea compound 3g | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-hydroxy-3-({[(pyridin-4-yl)methyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ... | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FQY
 
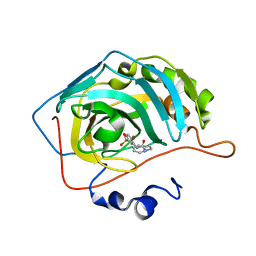 | | Carbonic Anhydrase II in complex with the alkyl urea 3h | | Descriptor: | 4-hydroxy-3-({[2-(pyridin-2-yl)ethyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.C, McKenna, R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | New Insights into Conformationally Restricted Carbonic Anhydrase Inhibitors.
Molecules, 28, 2023
|
|
8FR4
 
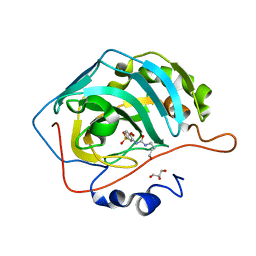 | |
8FR2
 
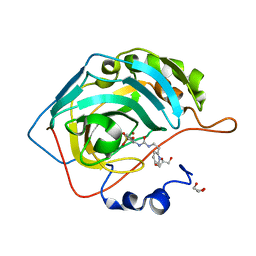 | |
5ZXI
 
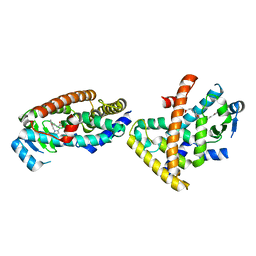 | | Co-crystal structure of an Inhibitor in complex with human PPARdelta LBD | | Descriptor: | 6-[2-({2-[4-(furan-2-yl)phenyl]-5-methyl-1H-imidazol-1-yl}methyl)phenoxy]hexanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Rani, S.T, Laxminarasimhan, A, Senaiar, R.S, Krishnamurthy, N. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective PPAR delta Modulators Improve Mitochondrial Function: Potential Treatment for Duchenne Muscular Dystrophy (DMD).
ACS Med Chem Lett, 9, 2018
|
|
5XMQ
 
 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and MF011 | | Descriptor: | 4-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]-~{N}-methyl-~{N}-(phenylmethyl)benzenesulfonamide, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5X02
 
 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor FF-10101 | | Descriptor: | N-[(2S)-1-[5-[2-[(4-cyanophenyl)amino]-4-(propylamino)pyrimidin-5-yl]pent-4-ynylamino]-1-oxidanylidene-propan-2-yl]-4-(dimethylamino)-N-methyl-but-2-enamide, Receptor-type tyrosine-protein kinase FLT3, SULFATE ION | | Authors: | Fujikawa, N, Hirano, D, Takasaki, M, Terada, D, Hagiwara, S, Park, S.-Y, Sugiyama, K. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells withFLT3mutations.
Blood, 131, 2018
|
|
5WOV
 
 | |
5WOW
 
 | |
5X8H
 
 | | Crystal structure of the ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 | | Descriptor: | Short-chain dehydrogenase reductase | | Authors: | Zhao, F.J, Jin, Y, Liu, Z.C, Wang, G.G, Wu, Z.L. | | Deposit date: | 2017-03-02 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and iterative saturation mutagenesis of ChKRED20 for expanded catalytic scope
Appl. Microbiol. Biotechnol., 101, 2017
|
|
5XMP
 
 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and MF057 | | Descriptor: | 4-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]-~{N}-methyl-~{N}-[2,2,2-tris(fluoranyl)ethyl]benzenesulfonamide, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5XMS
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS498 | | Descriptor: | (4~{S})-6-azanyl-4-[3-(2-fluorophenyl)-5-(trifluoromethyl)phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5- carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5XP5
 
 | | C-Src in complex with ATP-Chf | | Descriptor: | MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[(S)-fluoranyl-[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Duan, Y, Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
5XDJ
 
 | | Esculentin-1a(1-21)NH2 | | Descriptor: | Esculentin-1A | | Authors: | Ghosh, A, Bhunia, A. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Membrane perturbing activities and structural properties of the frog-skin derived peptide Esculentin-1a(1-21)NH2 and its Diastereomer Esc(1-21)-1c: Correlation with their antipseudomonal and cytotoxic activity
Biochim. Biophys. Acta, 1859, 2017
|
|
6ABZ
 
 | | Crystal Structure of HEWL in deionized water | | Descriptor: | CHLORIDE ION, Lysozyme C, S-1,2-PROPANEDIOL, ... | | Authors: | Seyedarabi, A, Seraj, Z. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of Cinnamaldehyde and Phenyl ethyl alcohol as two types of precipitants affecting protein hydration levels.
Int.J.Biol.Macromol., 146, 2019
|
|
6AMM
 
 | | CAT192 Fab Insertion Mutant H0/L1 | | Descriptor: | CAT192 Fab Heavy chain, CAT192 Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based engineering to restore high affinity binding of an isoform-selective anti-TGF beta 1 antibody.
MAbs, 10, 2018
|
|
6AGR
 
 | | Structure of HEWL co-crystallised with phenylethyl alcohol | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, ACETATE ION, ... | | Authors: | Seyedarabi, A, Seraj, Z. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The role of Cinnamaldehyde and Phenyl ethyl alcohol as two types of precipitants affecting protein hydration levels.
Int.J.Biol.Macromol., 146, 2019
|
|
5ZTN
 
 | | The crystal structure of human DYRK2 in complex with Curcumin | | Descriptor: | (1Z,4Z,6E)-5-hydroxy-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,4,6-trien-3-one, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Ji, C.G, Xiao, J.Y. | | Deposit date: | 2018-05-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
