8E81
 
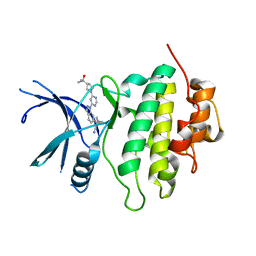 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 25 | | Descriptor: | (1S)-1-[(1P)-1-{6-[(3R)-3-(2-hydroxypropan-2-yl)pyrrolidin-1-yl]pyrimidin-4-yl}-1H-indazol-6-yl]spiro[2.2]pentane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
5Y5S
 
 | | Structure of photosynthetic LH1-RC super-complex at 1.9 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Yu, L.-J, Suga, M, Wang-Otomo, Z.-Y, Shen, J.-R. | | Deposit date: | 2017-08-09 | | Release date: | 2018-04-11 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of photosynthetic LH1-RC supercomplex at 1.9 angstrom resolution.
Nature, 556, 2018
|
|
4YOB
 
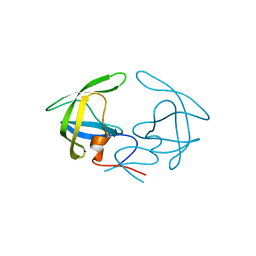 | | Crystal Structure of Apo HIV-1 Protease MDR769 L33F | | Descriptor: | HIV-1 Protease | | Authors: | Kuiper, B.D, Keusch, B, Dewdney, T.G, Chordia, P, Ross, K, Brunzelle, J.S, Kovari, I.A, MacArthur, R, Salimnia, H, Kovari, L.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | The L33F darunavir resistance mutation acts as a molecular anchor reducing the flexibility of the HIV-1 protease 30s and 80s loops.
Biochem Biophys Rep, 2, 2015
|
|
4Y6P
 
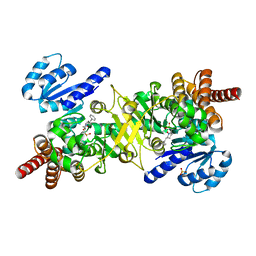 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, RC177, and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, CALCIUM ION, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2015-02-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Bioactivity of beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase.
J.Med.Chem., 58, 2015
|
|
4YL1
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 5-(4-tert-butylphenyl)-1-[4-(propan-2-yloxy)phenyl]-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
9EGO
 
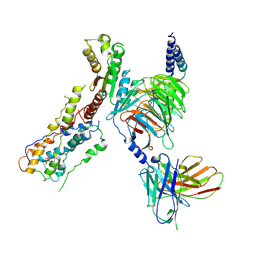 | | Cannabinoid receptor 1-Gi complex with novel ligand | | Descriptor: | Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tummino, T.A, Iliopoulos-Tsoutsouvas, C, Braz, J.M, O'Brien, E.S, Krishna Kumar, K, Makriyannis, M, Basbaum, A.I, Shoichet, B.K. | | Deposit date: | 2024-11-21 | | Release date: | 2025-04-30 | | Last modified: | 2025-09-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virtual library docking for cannabinoid-1 receptor agonists with reduced side effects.
Nat Commun, 16, 2025
|
|
5H9A
 
 | | Crystal structure of the Apo form of human cellular retinol binding protein 1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
3E10
 
 | |
6SEX
 
 | | X-ray structure of the gold/lysozyme adduct formed upon 21h exposure of protein crystals to compound 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ferraro, G, Giorgio, A, Merlino, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Protein-mediated disproportionation of Au(i): insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme.
Dalton Trans, 48, 2019
|
|
5NRB
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-1 | | Descriptor: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, N-acetyllactosaminide alpha-1,3-galactosyltransferase, ... | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6F1X
 
 | | Complex between MTH1 and compound 7 (a 7-azaindole-2-amide derivative) | | Descriptor: | 4-(3-chlorophenyl)-~{N}-ethyl-1~{H}-pyrrolo[2,3-b]pyridine-2-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
9DN3
 
 | |
4ZYI
 
 | |
6UDY
 
 | | X-ray co-crystal structure of compound 5 with Mcl-1 | | Descriptor: | (3S)-6'-chloro-5-(cyclobutylmethyl)-3',4,4',5-tetrahydro-2H,2'H-spiro[1,5-benzoxazepine-3,1'-naphthalene]-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
7N08
 
 | |
5C1Q
 
 | | Serine/threonine-protein kinase pim-1 | | Descriptor: | 3-methoxy[1]benzothieno[2,3-c]quinolin-6(5H)-one, Serine/threonine-protein kinase pim-1 | | Authors: | Li, W, Wan, X, Huang, N. | | Deposit date: | 2015-06-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Serine/threonine-protein kinase pim-1
To Be Published
|
|
3FSE
 
 | |
6UM8
 
 | | HIV Integrase in complex with Compound-14 | | Descriptor: | (2S)-tert-butoxy[7-(8-fluoro-5-methyl-3,4-dihydro-2H-1-benzopyran-6-yl)-5-methyl-2-phenylpyrazolo[1,5-a]pyrimidin-6-yl]acetic acid, DI(HYDROXYETHYL)ETHER, Integrase, ... | | Authors: | Khan, J.A, Kish, K. | | Deposit date: | 2019-10-09 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Optimization of Novel Pyrazolopyrimidines as Potent and Orally Bioavailable Allosteric HIV-1 Integrase Inhibitors.
J.Med.Chem., 63, 2020
|
|
5HHW
 
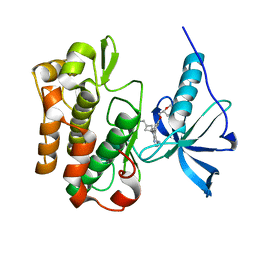 | | Crystal structure of insulin receptor kinase domain in complex with cis-(R)-7-(3-(azetidin-1-ylmethyl)cyclobutyl)-5-(3-((tetrahydro-2H-pyran-2-yl)methoxy)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine. | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-(azetidin-1-ylmethyl)cyclobutyl]-5-[3-[[(2~{R})-oxan-2-yl]methoxy]phenyl]pyrrolo[2,3-d]pyrimidin-4-amine, Insulin receptor | | Authors: | Scheufler, C, Izaac, A, Stauffer, F. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a 5-[3-phenyl-(2-cyclic-ether)-methylether]-4-aminopyrrolo[2,3-d]pyrimidine series of IGF-1R inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
9EB6
 
 | | Chicken YF1.7*1 presenting myristoylated peptide derived from tegument protein CIRC | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khandokar, Y, Wang, C.J.H, Rossjohn, J, Le Nours, J. | | Deposit date: | 2024-11-11 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for presentation of N-myristoylated peptides by the chicken YF1∗7.1 molecule.
J.Biol.Chem., 301, 2025
|
|
6UDX
 
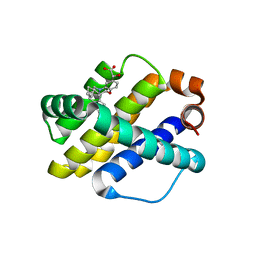 | | X-ray co-crystal structure of compound 7 with Mcl-1 | | Descriptor: | (2R)-[(3S)-6'-chloro-5-(cyclobutylmethyl)-3',4,4',5-tetrahydro-2H,2'H-spiro[1,5-benzoxazepine-3,1'-naphthalen]-7-yl](hydroxy)acetic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6UKI
 
 | |
5AAU
 
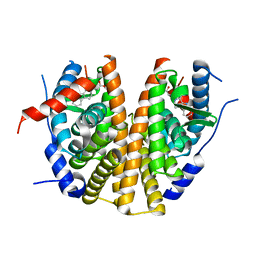 | | Optimization of a novel binding motif to to (E)-3-(3,5-difluoro-4-((1R,3R)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H- pyrido(3,4-b)indol-1-yl)phenyl)acrylic acid (AZD9496), a potent and orally bioavailable selective estrogen receptor downregulator and antagonist | | Descriptor: | 3-(1-(4-Chlorophenyl)-3,4-dihydro-1H-pyrido(3,4-b)indol-2(9H)-yl)propanoic acid, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Bradbury, R.H, de Almeida, C, Andrews, D.M, Ballard, P, Buttar, D, Callis, R.J, Currie, G.S, Curwen, J.O, Davies, C.D, de Savi, C, Donald, C.S, Feron, L.J.L, Glossop, S.C, Hayter, B.R, Karoutchi, G, Lamont, S.G, MacFaul, P, Moss, T, Pearson, S.E, Rabow, A.A, Tonge, M, Walker, G.E, Weir, H.M, Wilson, Z. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
3UZP
 
 | | crystal structure of ck1d with PF670462 from P21 crystal form | | Descriptor: | 4-[1-cyclohexyl-4-(4-fluorophenyl)-1H-imidazol-5-yl]pyrimidin-2-amine, Casein kinase I isoform delta | | Authors: | Huang, X. | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
6CZX
 
 | | Crystal structure of Arabidopsis thaliana phosphoserine aminotransferase isoform 1 (AtPSAT1) in complex with PLP internal aldimine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, Phosphoserine aminotransferase 1, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2018-04-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis of Phosphoserine Aminotransferase (Isoform 1) FromArabidopsis thaliana- the Enzyme Involved in the Phosphorylated Pathway of Serine Biosynthesis.
Front Plant Sci, 9, 2018
|
|
