1DP0
 
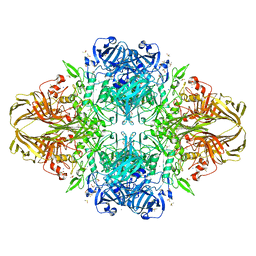 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | Descriptor: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1999-12-22 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
9MO0
 
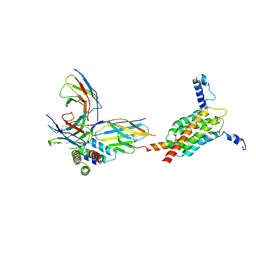 | | Cryo-EM structure of human MPC in complex with AKOS005153046 | | Descriptor: | (~{E})-2-cyano-3-[5-(2-nitrophenyl)furan-2-yl]prop-2-enoic acid, Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, ... | | Authors: | Zhang, J, He, Z, Feng, L. | | Deposit date: | 2024-12-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of mitochondrial pyruvate carrier and its inhibition mechanism.
Nature, 641, 2025
|
|
9MNZ
 
 | | Cryo-EM structure of human MPC in complex with UK5099 in nanodiscs | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, ... | | Authors: | Zhang, J, He, Z, Feng, L. | | Deposit date: | 2024-12-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of mitochondrial pyruvate carrier and its inhibition mechanism.
Nature, 641, 2025
|
|
9MNX
 
 | | Cryo-EM structure of human MPC in complex with UK5099 in LMNG | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, ... | | Authors: | Zhang, J, He, Z, Feng, L. | | Deposit date: | 2024-12-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of mitochondrial pyruvate carrier and its inhibition mechanism.
Nature, 641, 2025
|
|
1EDH
 
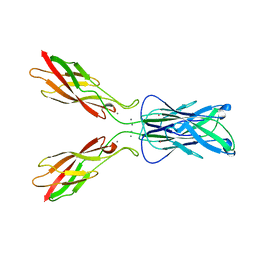 | | E-CADHERIN DOMAINS 1 AND 2 IN COMPLEX WITH CALCIUM | | Descriptor: | CALCIUM ION, E-CADHERIN, MERCURY (II) ION | | Authors: | Nagar, B, Overduin, M, Ikura, M, Rini, J.M. | | Deposit date: | 1996-05-15 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of calcium-induced E-cadherin rigidification and dimerization.
Nature, 380, 1996
|
|
8GJ3
 
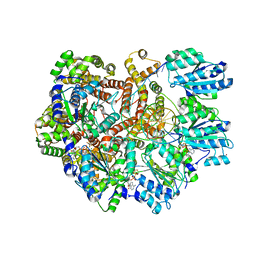 | | E. coli clamp loader on primed template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GJ1
 
 | | E. coli clamp loader with open clamp on primed template DNA (form 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta sliding clamp, DNA polymerase III subunit delta, ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GIY
 
 | | E. coli clamp loader with closed clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GJ0
 
 | | E. coli clamp loader with open clamp on primed template DNA (form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta sliding clamp, DNA polymerase III subunit delta, ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GIZ
 
 | | E. coli clamp loader with open clamp | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8GJ2
 
 | | E. coli clamp loader with closed clamp on primed template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta sliding clamp, DNA polymerase III subunit delta, ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli.
Nat Commun, 15, 2024
|
|
8F6C
 
 | | E. coli cytochrome bo3 ubiquinol oxidase dimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
8F68
 
 | | E. coli cytochrome bo3 ubiquinol oxidase monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A. | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
5J4D
 
 | |
6Y5M
 
 | | Crystal structure of mouse Autotaxin in complex with compound 1a | | Descriptor: | (~{E})-3-[4-chloranyl-2-[(5-methyl-1,2,3,4-tetrazol-2-yl)methyl]phenyl]-1-[(2~{R})-4-[(4-fluorophenyl)methyl]-2-methyl-piperazin-1-yl]prop-2-en-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Development of autotaxin inhibitors: A series of tetrazole cinnamides.
Bioorg.Med.Chem.Lett., 31, 2021
|
|
5LRY
 
 | | E coli [NiFe] Hydrogenase Hyd-1 mutant E28D | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J.Am.Chem.Soc., 140, 2018
|
|
5LJO
 
 | | E. coli BAM complex (BamABCDE) by cryoEM | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Iadanza, M.G, Ranson, N.A, Radford, S.E, Higgins, A.J, Schffrin, B, Calabrese, A.N, Ashcroft, A.E, Brockwell, D.J. | | Deposit date: | 2016-07-19 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM.
Nat Commun, 7, 2016
|
|
9FL3
 
 | | Crystal structure of IL-17A in complex with compound 26 | | Descriptor: | (~{E})-~{N}-[(~{S})-[4,4-bis(fluoranyl)cyclohexyl]-[7-[(1~{S})-2-methoxy-1-[(4~{S})-2-oxidanylidene-4-(trifluoromethyl)imidazolidin-1-yl]ethyl]imidazo[1,2-b]pyridazin-2-yl]methyl]-3-cyclopropyl-2-fluoranyl-prop-2-enamide, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Discovery and In Vivo Exploration of 1,3,4-Oxadiazole and alpha-Fluoroacrylate Containing IL-17 Inhibitors.
J.Med.Chem., 67, 2024
|
|
6QEM
 
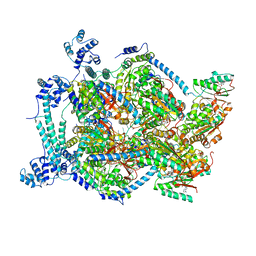 | | E. coli DnaBC complex bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein DnaC, MAGNESIUM ION, ... | | Authors: | Arias-Palomo, E, Puri, N, O'Shea Murray, V.L, Yan, Q, Berger, J.M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.
Mol.Cell, 74, 2019
|
|
6QEL
 
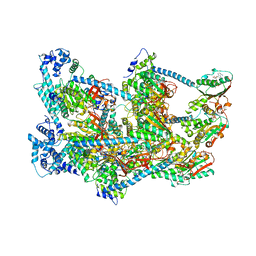 | | E. coli DnaBC apo complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein dnaC, MAGNESIUM ION, ... | | Authors: | Arias-Palomo, E, Puri, N, O'Shea Murray, V.L, Yan, Q, Berger, J.M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.
Mol.Cell, 74, 2019
|
|
9CS0
 
 | | E. coli BamA beta-barrel bound to darobactin and cyclic peptide CP1 | | Descriptor: | Cyclic peptide CP1, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
9CS1
 
 | | E. coli BamA beta-barrel bound to darobactin and cyclic peptide CP2 | | Descriptor: | Cyclic peptide CP2, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
9CS2
 
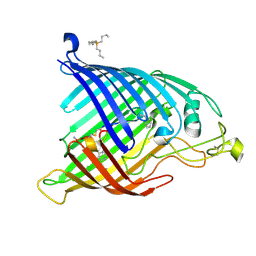 | | E. coli BamA beta-barrel bound to cyclic peptide CP3 | | Descriptor: | Cyclic peptide CP3, Outer membrane protein assembly factor BamA, tetrabutylphosphonium | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
5WSN
 
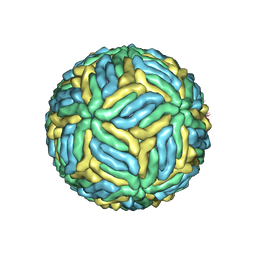 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
5MK4
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 7 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-5-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Andrei, S.A, Scheepstra, M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2016-12-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
