8UNJ
 
 | | Atomic model of the human CTF18-RFC alone in the apo state (State 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UMT
 
 | | Atomic model of the human CTF18-RFC-PCNA binary complex in the three-subunit binding state (state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UN0
 
 | | Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with cracked and closed PCNA (state 7) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, DNA (20-MER), ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UMU
 
 | | Atomic model of the human CTF18-RFC-PCNA binary complex in the four-subunit binding state (state 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UMY
 
 | | Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state II (state 6) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, DNA (20-MER), ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UMW
 
 | | Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex in the five-subunit binding state (state 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, DNA (20-MER), ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UMV
 
 | | Atomic model of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state I (state 5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, DNA (20-MER), ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OKA
 
 | | Human Mitochondrial Lon Y394F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OM7
 
 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OJL
 
 | | Human Mitochondrial Lon Y394E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8RHN
 
 | | Structure of the 55LCC ATPase complex | | Descriptor: | ATPase family gene 2 protein homolog A, ATPase family gene 2 protein homolog B, Cyclin-dependent kinase 2-interacting protein, ... | | Authors: | Foglizzo, M, Degtjarik, O, Zeqiraj, E. | | Deposit date: | 2023-12-15 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
8XON
 
 | |
8XOO
 
 | |
8OOI
 
 | | Full composite cryo-EM map of p97/VCP in ADP.Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheng, T.C, Sakata, E, Schuetz, A.K. | | Deposit date: | 2023-04-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Characterizing ATP processing by the AAA+ protein p97 at the atomic level.
Nat.Chem., 16, 2024
|
|
8HRZ
 
 | |
8UNH
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UK9
 
 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U0V
 
 | | S. cerevisiae Pex1/Pex6 with 1 mM ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peroxisomal ATPase PEX1, Peroxisomal ATPase PEX6 | | Authors: | Gardner, B.M. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | The N1 domain of the peroxisomal AAA-ATPase Pex6 is required for Pex15 binding and proper assembly with Pex1.
J.Biol.Chem., 300, 2023
|
|
8UH7
 
 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | Authors: | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.628 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
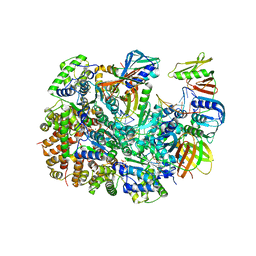 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8K3Y
 
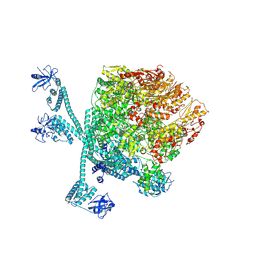 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPJ
 
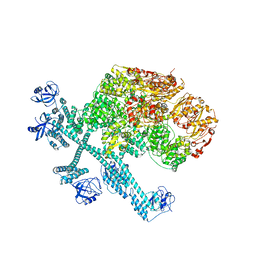 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUU
 
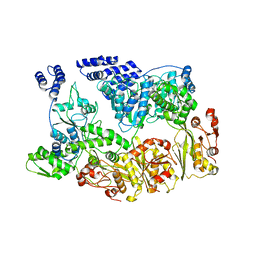 | | MtaLon-ADP for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUV
 
 | | MtaLon-ADP for the spiral oligomers of tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUW
 
 | | MtaLon-ADP for the spiral oligomers of pentamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
