8PN1
 
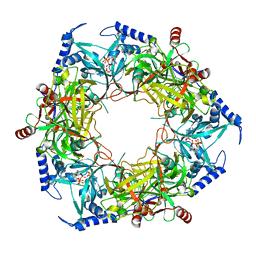 | | CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMM
 
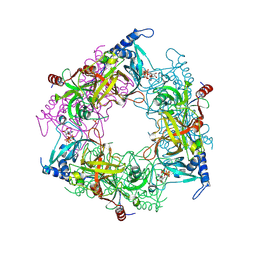 | | Structure of Nal1 protein, allele SPIKE from japonica rice, construct 31-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PML
 
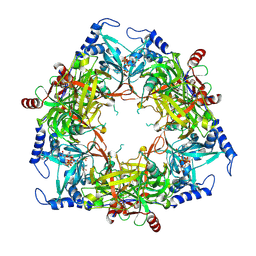 | | Structure of Nal1 protein , SPIKE allele from japonica rice, construct 46-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMI
 
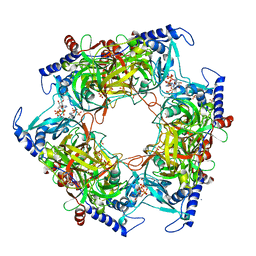 | | Structure of Nal1 indica cultivar IR64, construct 36-458 in presence of peptide from FZP protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AMMONIUM ION, MAGNESIUM ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMG
 
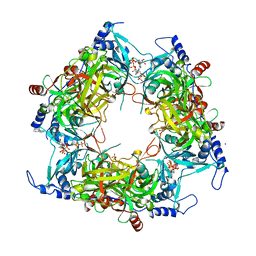 | | Structure of Nal1 indica cultivar IR64, construct 36-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8OVY
 
 | | Structure of analogue of superfolded GFP | | Descriptor: | Green fluorescent protein | | Authors: | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
8OVP
 
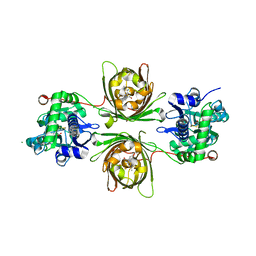 | | X-ray structure of the iAspSnFR in complex with L-aspartate | | Descriptor: | ACETATE ION, ASPARTIC ACID, MAGNESIUM ION, ... | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iAspSnFR in complex with L-aspartate
To Be Published
|
|
8OVO
 
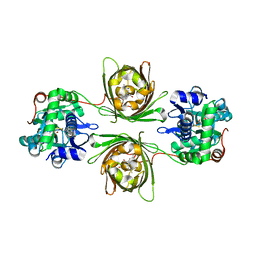 | | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A in complex with L-aspartate
To Be Published
|
|
8OVN
 
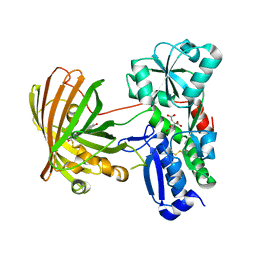 | | X-ray structure of the SF-iGluSnFR-S72A | | Descriptor: | CITRIC ACID, Putative periplasmic binding transport protein,Green fluorescent protein | | Authors: | Tarnawski, M, Hellweg, L, Bergner, A, Hiblot, J, Leippe, P, Johnsson, K. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the SF-iGluSnFR-S72A
To Be Published
|
|
8OTS
 
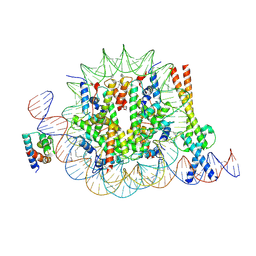 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8JZU
 
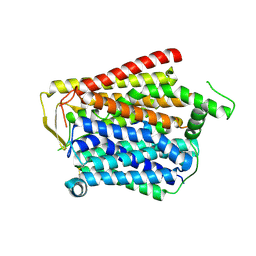 | | SLC15A4_TASL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TLR adapter,Green fluorescent protein, TSLAA-EGPF tag fusion protein | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
8JVJ
 
 | | Structure of human TRPV4 with antagonist A2 and RhoA | | Descriptor: | Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 2024
|
|
8JVI
 
 | | Structure of human TRPV4 with antagonist A2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 2024
|
|
8JU6
 
 | | Structure of human TRPV4 with antagonist GSK279 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 2024
|
|
8JU5
 
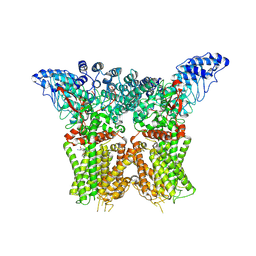 | | Structure of human TRPV4 with antagonist A1 | | Descriptor: | 4-[(3~{S},4~{S})-4-(aminomethyl)-1-(5-chloranylpyridin-2-yl)sulfonyl-4-oxidanyl-pyrrolidin-3-yl]oxy-2-fluoranyl-benzenecarbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 2024
|
|
8J7M
 
 | | ion channel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, ion channel,Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment) | | Authors: | Chen, H.W, Chen, H.W. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | ion channel
To Be Published
|
|
8J7F
 
 | | ion channel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Chen, H.W, Chen, H.W. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | structure of ion channel
To Be Published
|
|
8J6O
 
 | | transport T2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein (Fragment),SID1 transmembrane family member 2, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 2024
|
|
8J6M
 
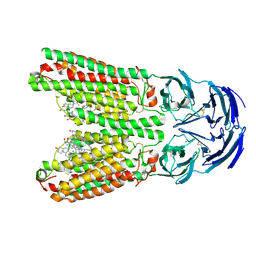 | | SIDT1 protein | | Descriptor: | CHOLESTEROL, Green fluorescent protein,SID1 transmembrane family member 1, OLEIC ACID, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 2024
|
|
8J1E
 
 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8IMY
 
 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, T, Xu, Y, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | Descriptor: | Beta-amyloid, Split Green flourescent protein | | Authors: | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
8I4K
 
 | | Structure of Azami Red1.0, a red fluorescent protein engineered from Azami Green | | Descriptor: | Azami Red1.0, CALCIUM ION | | Authors: | Otsubo, S, Takekawa, N, Imamura, H, Imada, K. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Red fluorescent proteins engineered from green fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
