1KJ6
 
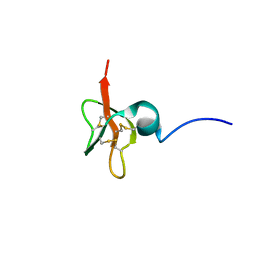 | | Solution Structure of Human beta-Defensin 3 | | Descriptor: | Beta-defensin 3 | | Authors: | Schibli, D.J, Hunter, H.N, Aseyev, V, Starner, T.D, Wiencek, J.M, McCray Jr, P.B, Tack, B.F, Vogel, H.J. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
J.Biol.Chem., 277, 2002
|
|
7AC4
 
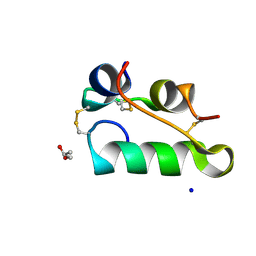 | | Structure of insulin collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | Insulin, R-1,2-PROPANEDIOL, SODIUM ION | | Authors: | Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Vera, L, Wang, M, Marsh, M. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1KJT
 
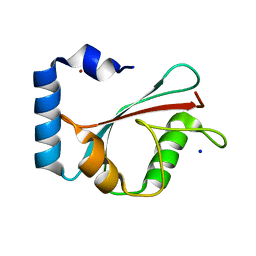 | | Crystal Structure of the GABA(A) Receptor Associated Protein, GABARAP | | Descriptor: | GABARAP, NICKEL (II) ION, SODIUM ION | | Authors: | Bavro, V.N, Sola, M, Bracher, A, Kneussel, M, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the GABA(A)-receptor-associated protein, GABARAP.
EMBO Rep., 3, 2002
|
|
9C41
 
 | |
7AC2
 
 | | Structure of Hen Egg White Lysozyme collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | CHLORIDE ION, Lysozyme, SODIUM ION | | Authors: | Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Vera, L, Wang, M, Marsh, M. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1KK0
 
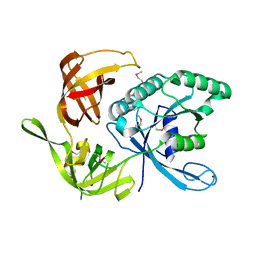 | |
9C40
 
 | |
7AC3
 
 | | Structure of thaumatin collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | L(+)-TARTARIC ACID, S-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Vera, L, Wang, M, Marsh, M. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1KAB
 
 | |
9C43
 
 | |
1KKB
 
 | | Complex of Escherichia coli Adenylosuccinate Synthetase with IMP and Hadacidin | | Descriptor: | Adenylosuccinate Synthetase, HADACIDIN, INOSINIC ACID | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-06 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
9C7S
 
 | |
7ABW
 
 | | Crystal structure of siderophore reductase FoxB | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DECYL-BETA-D-MALTOPYRANOSIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural insights into a novel family of integral membrane siderophore reductases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1KO0
 
 | | Crystal Structure of a D,L-lysine complex of diaminopimelate decarboxylase | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, LYSINE, ... | | Authors: | Levdikov, V, Blagova, L, Bose, N, Momany, C. | | Deposit date: | 2001-12-19 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diaminopimelate Decarboxylase uses a Versatile Active Site for Stereospecific Decarboxylation
To be Published
|
|
7AC5
 
 | | Structure of Tubulin Darpin complex 1 collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Martiel, I, Olieric, N, Wranik, M, Padeste, C, Karpik, A, Huang, C.Y, Wang, M, Marsh, M. | | Deposit date: | 2020-09-10 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1KOG
 
 | | Crystal structure of E. coli threonyl-tRNA synthetase interacting with the essential domain of its mRNA operator | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase, Threonyl-tRNA synthetase mRNA, ... | | Authors: | Torres-Larrios, A, Dock-Bregeon, A.C, Romby, P, Rees, B, Sankaranarayanan, R, Caillet, J, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
Nat.Struct.Biol., 9, 2002
|
|
9C3R
 
 | |
1KKM
 
 | | L.casei HprK/P in complex with B.subtilis P-Ser-HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHATE ION, ... | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7AEQ
 
 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-4-(2-hydroxyethylsulfanyl)-N-methyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-4-(2-hydroxyethylsulfanyl)-~{N}-methyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
9C6Z
 
 | | Crystal structure of Staphylococcal Superantigen-Like protein 11 in complex with FB127 | | Descriptor: | (4aR,5S,6S,8R,8aS)-6-(hydroxymethyl)-8-methoxy-2,2-dimethylhexahydro-2H-pyrano[3,4-d][1,3]dioxin-5-ol, N-acetyl-alpha-neuraminic acid, PENTAETHYLENE GLYCOL, ... | | Authors: | Finton, S, Birrane, G. | | Deposit date: | 2024-06-10 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structure of FB127 in Complex with SSL11
To Be Published
|
|
1KL4
 
 | |
7AES
 
 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-N-methyl-4-propylsulfanyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-~{N}-methyl-4-propylsulfanyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICINE, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
1KQL
 
 | | Crystal structure of the C-terminal region of striated muscle alpha-tropomyosin at 2.7 angstrom resolution | | Descriptor: | Fusion Protein of and striated muscle alpha-tropomyosin and the GCN4 leucine zipper | | Authors: | Li, Y, Mui, S, Brown, J.H, Strand, J, Reshetnikova, L, Tobacman, L.S, Cohen, C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQO
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
9C3Z
 
 | |
