1HW9
 
 | |
2FWV
 
 | | Crystal Structure of Rv0813 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, hypothetical protein MtubF_01000852 | | Authors: | Shepard, W, Haouz, A, Grana, M, Buschiazzo, A, Betton, J.M, Cole, S.T, Alzari, P.M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-03 | | Release date: | 2006-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Rv0813c from Mycobacterium tuberculosis Reveals a New Family of Fatty Acid-Binding Protein-Like Proteins in Bacteria
J.Bacteriol., 189, 2007
|
|
6YUA
 
 | |
6YTT
 
 | | CO-dehydrogenase/Acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum at 3.0-A resolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CO dehydrogenase/acetyl-CoA synthase complex, beta subunit, ... | | Authors: | Wagner, T, Lemaire, O.N. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Gas channel rerouting in a primordial enzyme: Structural insights of the carbon-monoxide dehydrogenase/acetyl-CoA synthase complex from the acetogen Clostridium autoethanogenum.
Biochim Biophys Acta Bioenerg, 1862, 2020
|
|
5OOL
 
 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5CUF
 
 | | X-ray crystal structure of SeMet human Sestrin2 | | Descriptor: | Sestrin-2 | | Authors: | Kim, H, An, S, Ro, S.-H, Lee, J.H, Cho, U.-S. | | Deposit date: | 2015-07-24 | | Release date: | 2016-01-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains.
Nat Commun, 6, 2015
|
|
6ZI7
 
 | | Crystal structure of OleP-oleandolide(DEO) bound to L-rhamnose | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI3
 
 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZHZ
 
 | | OleP-oleandolide(DEO) in high salt crystallization conditions | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Cecchetti, C. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI2
 
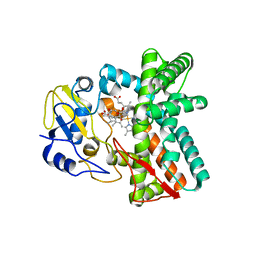 | | OleP-oleandolide(DEO) in low salt crystallization conditions | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Parisi, G, Cecchetti, C. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
5DIQ
 
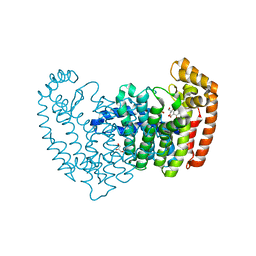 | | Crystal structure of human FPPS in complex with salicylic acid derivative 3a | | Descriptor: | 2-(naphthalen-1-ylmethoxy)benzoic acid, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Rondeau, J.M, Bourgier, E, Lehmann, S. | | Deposit date: | 2015-09-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel Allosteric Non-Bisphosphonate Inhibitors of Farnesyl Pyrophosphate Synthase by Integrated Lead Finding.
Chemmedchem, 10, 2015
|
|
5DJP
 
 | |
5DJV
 
 | |
5DJR
 
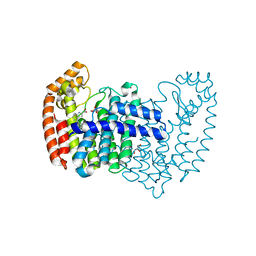 | | Crystal structure of human FPPS in complex with biaryl compound 6 | | Descriptor: | 1H,1'H-4,4'-biindole-2-carboxylic acid, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Rondeau, J.M, Bourgier, E, Lehmann, S. | | Deposit date: | 2015-09-02 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Novel Allosteric Non-Bisphosphonate Inhibitors of Farnesyl Pyrophosphate Synthase by Integrated Lead Finding.
Chemmedchem, 10, 2015
|
|
2MDI
 
 | | Solution structure of the PP2WW mutant (KPP2WW) of HYPB | | Descriptor: | Histone-lysine N-methyltransferase SETD2 | | Authors: | Gao, Y.G, Hu, H.Y. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin.
Structure, 22, 2014
|
|
2LXL
 
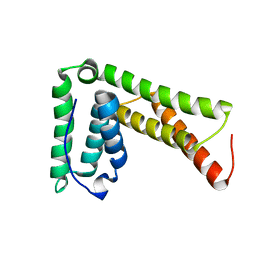 | | Lip5(mit)2 | | Descriptor: | Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions of the Human LIP5 Regulatory Protein with Endosomal Sorting Complexes Required for Transport.
J.Biol.Chem., 287, 2012
|
|
2LXM
 
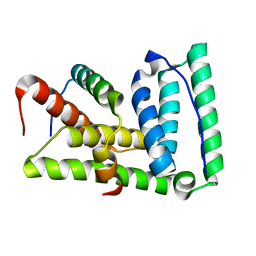 | | Lip5-chmp5 | | Descriptor: | Charged multivesicular body protein 5, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions of the Human LIP5 Regulatory Protein with Endosomal Sorting Complexes Required for Transport.
J.Biol.Chem., 287, 2012
|
|
2MDJ
 
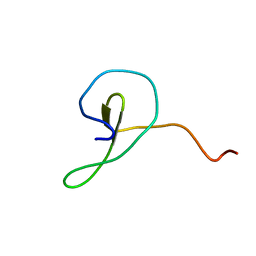 | |
2MDC
 
 | | Solution structure of the WW domain of HYPB | | Descriptor: | Histone-lysine N-methyltransferase SETD2 | | Authors: | Gao, Y.G. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin
Structure, 22, 2014
|
|
6J8Y
 
 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
2N1O
 
 | |
2NZ0
 
 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
2Q5A
 
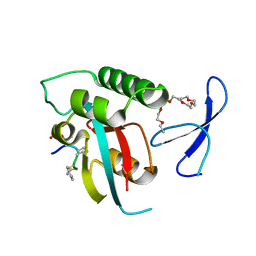 | | human Pin1 bound to L-PEPTIDE | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
2FIC
 
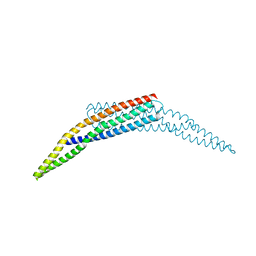 | | The crystal structure of the BAR domain from human Bin1/Amphiphysin II and its implications for molecular recognition | | Descriptor: | Myc box-dependent-interacting protein 1, XENON | | Authors: | Casal, E, Federici, L, Zhang, W, Fernandez-Recio, J, Priego, E.M, Miguel, R.N, Duhadaway, J.B, Prendergast, G.C, Luisi, B.F, Laue, E.D. | | Deposit date: | 2005-12-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crystal Structure of the BAR Domain from Human Bin1/Amphiphysin II and Its Implications for Molecular Recognition
Biochemistry, 45, 2006
|
|
6ZE5
 
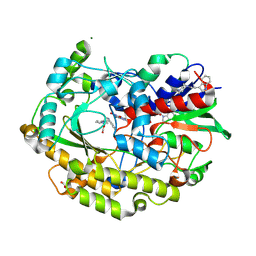 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
