4XFY
 
 | |
3DB7
 
 | |
3DEH
 
 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, isoquinoline-1,3,4(2H)-trione | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
4XUO
 
 | |
6CW7
 
 | | E. coli DHFR product complex with (6S)-5,6,7,8-TETRAHYDROFOLATE | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-30 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
5AXR
 
 | |
5L0R
 
 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5F4N
 
 | | Multi-parameter lead optimization to give an oral CHK1 inhibitor clinical candidate: (R)-5-((4-((morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase Chk1, ... | | Authors: | Collins, I, Garrett, M.D, van Montfort, R, Osborne, J.D, Matthews, T.P, McHardy, T, Proisy, N, Cheung, K.J, Lainchbury, M, Brown, N, Walton, M.I, Eve, P.D, Boxall, K.J, Hayes, A, Henley, A.T, Valenti, M.R, De Haven Brandon, A.K, Box, G, Westwood, I.M, Jamin, Y, Robinson, S.P, Leonard, P, Reader, J.C, Aherne, G.W, Raynaud, F.I, Eccles, S.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Multiparameter Lead Optimization to Give an Oral Checkpoint Kinase 1 (CHK1) Inhibitor Clinical Candidate: (R)-5-((4-((Morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737).
J.Med.Chem., 59, 2016
|
|
3P02
 
 | |
6PNO
 
 | |
4XVT
 
 | | Crystal structure of HIV-1 93TH057 coreE gp120 with antibody 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) Light chain, ENVELOPE GLYCOPROTEIN GP120 OF HIV-1 CLADE A/E, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
6D9X
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | 2-phenyl-6,7-dihydro-5H-pyrrolo[1,2-a]imidazole, SODIUM ION, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
3DEK
 
 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, N-[3-(2-fluoroethoxy)phenyl]-N'-(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-6-yl)butanediamide | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
6SET
 
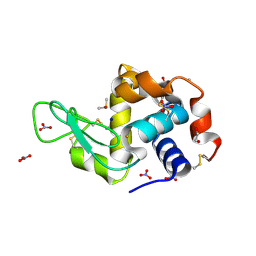 | | X-ray structure of the gold/lysozyme adduct formed upon 3 days exposure of protein crystals to compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ferraro, G, Giorgio, A, Merlino, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-mediated disproportionation of Au(i): insights from the structures of adducts of Au(iii) compounds bearing N,N-pyridylbenzimidazole derivatives with lysozyme.
Dalton Trans, 48, 2019
|
|
3DLK
 
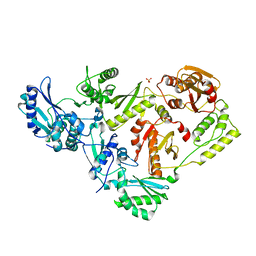 | | Crystal Structure of an engineered form of the HIV-1 Reverse Transcriptase, RT69A | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, p51 RT | | Authors: | Ho, W.C, Bauman, J.D, Himmel, D.M, Das, K, Arnold, E. | | Deposit date: | 2008-06-27 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design.
Nucleic Acids Res., 36, 2008
|
|
3DNL
 
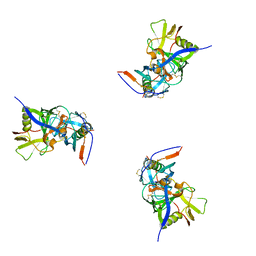 | | Molecular structure for the HIV-1 gp120 trimer in the b12-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
6DAR
 
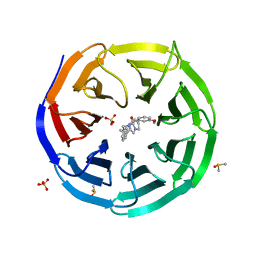 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | DIMETHYL SULFOXIDE, N-(cyclopropylmethyl)-N-{[3-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)phenyl]methyl}-3-methoxybenzamide, SULFATE ION, ... | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
2VPO
 
 | | High resolution structure of the periplasmic binding protein TeaA from TeaABC TRAP transporter of Halomonas elongata in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, MAGNESIUM ION, PERIPLASMIC SUBSTRATE BINDING PROTEIN | | Authors: | Kuhlmann, S.I, Terwisscha van Scheltinga, A.C, Bienert, R, Kunte, H.J, Ziegler, C. | | Deposit date: | 2008-03-03 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.55 A Structure of the Ectoine Binding Protein Teaa of the Osmoregulated Trap-Transporter Teaabc from Halomonas Elongata.
Biochemistry, 47, 2008
|
|
6YBK
 
 | | Structure of MBP-Mcl-1 in complex with compound 4d | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-(pyrazin-2-ylmethoxy)phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
4XUY
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger | | Descriptor: | GLYCEROL, Probable endo-1,4-beta-xylanase C, SULFATE ION | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.0027 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger
To Be Published
|
|
6SVP
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6Q8Z
 
 | | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide
To Be Published
|
|
3DM6
 
 | | Beta-secretase 1 complexed with statine-based inhibitor | | Descriptor: | 5-[[(2S)-2-[[(3R,4S)-5-(3,5-difluorophenoxy)-3-hydroxy-4-[[3-(methyl-methylsulfonyl-amino)-5-[[(1R)-1-phenylethyl]carbamoyl]phenyl]carbonylamino]pentanoyl]amino]-3-methyl-butanoyl]amino]benzene-1,3-dicarboxylic acid, Beta-secretase 1, ISOPROPYL ALCOHOL | | Authors: | Lindberg, J, Borkakoti, N, Nystrom, S. | | Deposit date: | 2008-06-30 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and SAR of potent statine-based BACE-1 inhibitors: exploration of P1 phenoxy and benzyloxy residues
Bioorg.Med.Chem., 16, 2008
|
|
3E02
 
 | |
6DAK
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | DIMETHYL SULFOXIDE, N-{[3-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)phenyl]methyl}benzamide, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
