3UC3
 
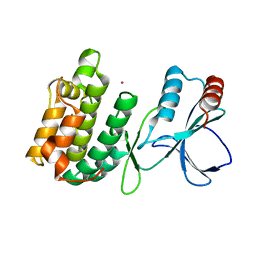 | | The crystal structure of Snf1-related kinase 2.3 | | Descriptor: | COBALT (II) ION, Serine/threonine-protein kinase SRK2I | | Authors: | Zhou, X.E, Ng, L.-M, Soon, F.-F, Kovach, A, Suino-Powell, K.M, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-10-25 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1VYP
 
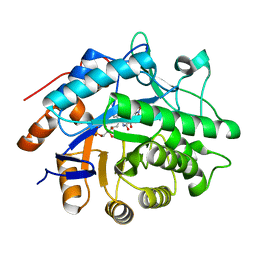 | | Structure of pentaerythritol tetranitrate reductase W102F mutant and complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
1MBO
 
 | |
5VJG
 
 | |
5VR2
 
 | |
5VSQ
 
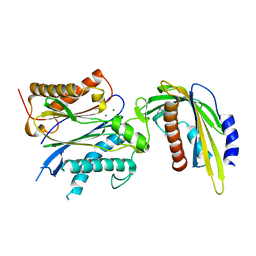 | | ABA-mimicking ligand AMF2beta in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(3,5-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.618 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
1MPJ
 
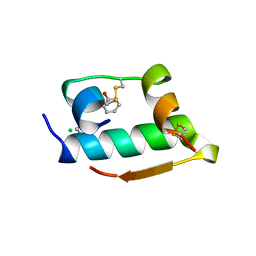 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | CHLORIDE ION, PHENOL, PHENOL INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1MTB
 
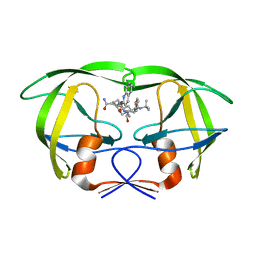 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
1MTN
 
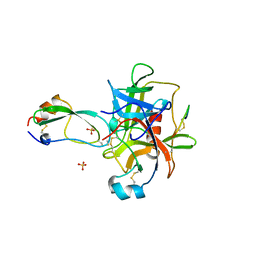 | | BOVINE ALPHA-CHYMOTRYPSIN:BPTI CRYSTALLIZATION | | Descriptor: | ALPHA-CHYMOTRYPSIN, BASIC PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Capasso, C, Rizzi, M, Menegatti, E, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the bovine alpha-chymotrypsin:Kunitz inhibitor complex. An example of multiple protein:protein recognition sites.
J.Mol.Recog., 10, 1997
|
|
1MT7
 
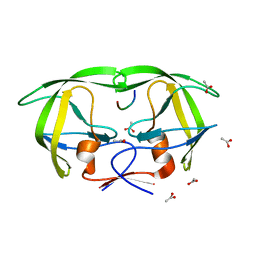 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | ACETATE ION, PROTEASE RETROPEPSIN, Substrate analogue | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
2FYM
 
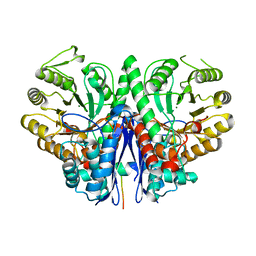 | |
4WAB
 
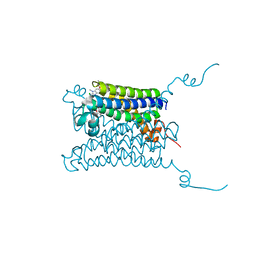 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
1N0N
 
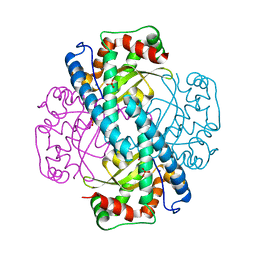 | | Catalytic and Structural Effects of Amino-Acid Substitution at His30 in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] | | Authors: | Borgstahl, G.E.O, Parge, H.E, Hickey, M.J, Beyer Jr, W.F, Hallewell, R.A, Tainer, J.A. | | Deposit date: | 2002-10-14 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Catalytic and Structural Effects of Amino-Acid Substitution at His30 in Human Manganese Superoxide Dismutase
to be published
|
|
1MWC
 
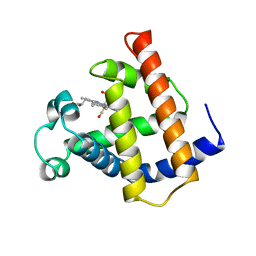 | | WILD TYPE MYOGLOBIN WITH CO | | Descriptor: | CARBON MONOXIDE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
4OH2
 
 | | Crystal Structure of Cu/Zn Superoxide Dismutase I149T | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Crane, B.R, Merz, G.E. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-15 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Copper-Based Pulsed Dipolar ESR Spectroscopy as a Probe of Protein Conformation Linked to Disease States.
Biophys.J., 107, 2014
|
|
7RDX
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDZ
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
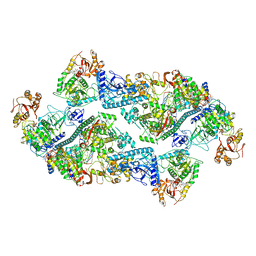 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
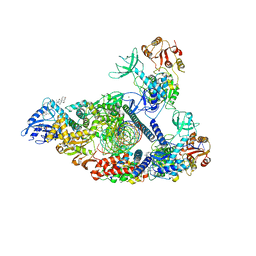 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
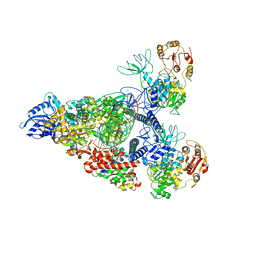 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
4FBY
 
 | | fs X-ray diffraction of Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Hellmich, J, Tran, R, Hattne, J, Laksmono, H, Gloeckner, C, Echols, N, Sierra, R.G, Sellberg, J, Lassalle-Kaiser, B, Gildea, R.J, Glatzel, P, Grosse-Kunstleve, R.W, Latimer, M.J, Mcqueen, T.A, Difiore, D, Fry, A.R, Messerschmidt, M.M, Miahnahri, A, Schafer, D.W, Seibert, M.M, Sokaras, D, Weng, T.-C, Zwart, P.H, White, W.E, Adams, P.D, Bogan, M.J, Boutet, S, Williams, G.J, Messinger, J, Sauter, N.K, Zouni, A, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (6.56 Å) | | Cite: | Room temperature femtosecond X-ray diffraction of photosystem II microcrystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V7I
 
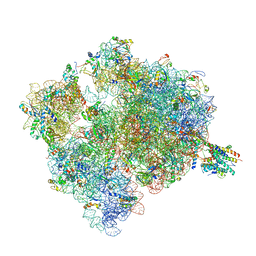 | | Ribosome-SecY complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gumbart, J.C, Trabuco, L.G, Schreiner, E, Villa, E, Schulten, K. | | Deposit date: | 2009-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Regulation of the protein-conducting channel by a bound ribosome.
Structure, 17, 2009
|
|
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
