2PNS
 
 | | 1.9 Angstrom resolution crystal structure of a plant cysteine protease Ervatamin-C refinement with cDNA derived amino acid sequence | | Descriptor: | Ervatamin-C, a papain-like plant cysteine protease, PHOSPHATE ION, ... | | Authors: | Ghosh, R, Guha Thakurta, P, Biswas, S, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A thermostable cysteine protease precursor from a tropical plant contains an unusual C-terminal propeptide: cDNA cloning, sequence comparison and molecular modeling studies.
Biochem.Biophys.Res.Commun., 362, 2007
|
|
2PNT
 
 | | Crystal structure of the PDZ domain of human GRASP (GRP1) in complex with the C-terminal peptide of the metabotropic glutamate receptor type 1 | | Descriptor: | CHLORIDE ION, General receptor for phosphoinositides 1-associated scaffold protein | | Authors: | Elkins, J, Papagrigoriou, E, Cooper, C, Gileadi, C, Uppenberg, J, Bray, J, von Delft, F, Pike, A.C.W, Ugochukwu, E, Umeano, C, Gileadi, O, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-25 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
2PNU
 
 | | Crystal structure of human androgen receptor ligand-binding domain in complex with EM-5744 | | Descriptor: | (5S,8R,9S,10S,13R,14S,17S)-13-{2-[(3,5-DIFLUOROBENZYL)OXY]ETHYL}-17-HYDROXY-10-METHYLHEXADECAHYDRO-3H-CYCLOPENTA[A]PHENANTHREN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Cantin, L, Faucher, F, Couture, J.F, Pereira de Jesus-Tran, K, Legrand, P, Ciobanu, C.L, Singh, S.M, Labrie, F, Breton, R. | | Deposit date: | 2007-04-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Androgen Receptor Ligand-binding Domain Complexed with EM5744, a Rationally Designed Steroidal Ligand Bearing a Bulky Chain Directed toward Helix 12.
J.Biol.Chem., 282, 2007
|
|
2PNV
 
 | | Crystal Structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SKCa) channel from Rattus norvegicus | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Kim, J.Y, Kim, M.K, Kang, G.B, Park, C.S, Eom, S.H. | | Deposit date: | 2007-04-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SK(Ca)) channel from Rattus norvegicus.
Proteins, 70, 2008
|
|
2PNW
 
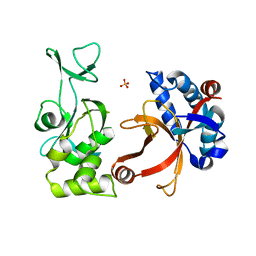 | | Crystal structure of membrane-bound lytic murein transglycosylase from Agrobacterium tumefaciens | | Descriptor: | Membrane-bound lytic murein transglycosylase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-25 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of membrane-bound lytic murein transglycosylase from Agrobacterium tumefaciens.
To be Published
|
|
2PNX
 
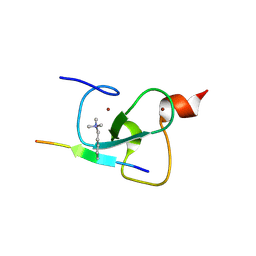 | |
2PNY
 
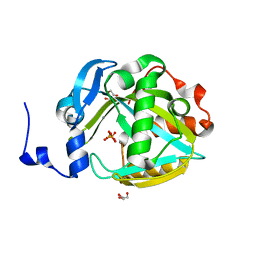 | | Structure of Human Isopentenyl-diphosphate Delta-isomerase 2 | | Descriptor: | CALCIUM ION, GLYCEROL, Isopentenyl-diphosphate Delta-isomerase 2, ... | | Authors: | Walker, J.R, Davis, T, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-25 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of the human Isopentenyl-diphosphate Delta-isomerase 2.
To be Published
|
|
2PNZ
 
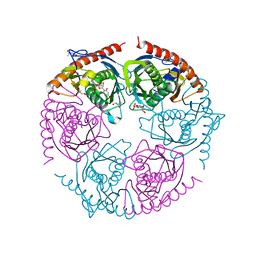 | |
2PO0
 
 | |
2PO1
 
 | |
2PO2
 
 | |
2PO3
 
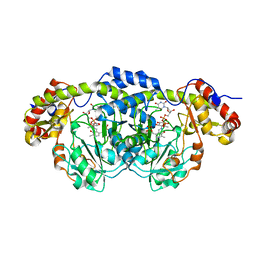 | | Crystal Structure Analysis of DesI in the presence of its TDP-sugar product | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 4-dehydrase | | Authors: | Burgie, E.S, Holden, H.M. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Architecture of DesI: A Key Enzyme in the Biosynthesis of Desosamine
Biochemistry, 46, 2007
|
|
2PO4
 
 | |
2PO5
 
 | | Crystal structure of human ferrochelatase mutant with His 263 replaced by Cys | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, A.E.W, Burden, A, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-25 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis
Biochemistry, 46, 2007
|
|
2PO6
 
 | |
2PO7
 
 | | Crystal structure of human ferrochelatase mutant with His 341 replaced by Cys | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis.
Biochemistry, 46, 2007
|
|
2PO8
 
 | |
2POA
 
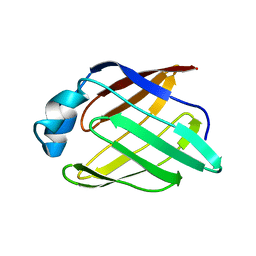 | | Schistosoma mansoni Sm14 Fatty Acid-Binding Protein: improvement of protein stability by substitution of the single Cys62 residue | | Descriptor: | 14 kDa fatty acid-binding protein | | Authors: | Ramos, C.R.R, Oyama Jr, S, Sforca, M.L, Pertinhez, T.A, Ho, P.L, Spisni, A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stability improvement of the fatty acid binding protein Sm14 from S. mansoni by Cys replacement: Structural and functional characterization of a vaccine candidate.
Biochim.Biophys.Acta, 1794, 2009
|
|
2POB
 
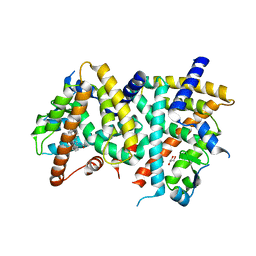 | | PPARgamma Ligand binding domain complexed with a farglitazar analogue gw4709 | | Descriptor: | GLYCEROL, N-[(2S)-2-[(2-BENZOYLPHENYL)AMINO]-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL]ACETAMIDE, Peroxisome proliferator-activated receptor gamma | | Authors: | Nolte, R.T. | | Deposit date: | 2007-04-26 | | Release date: | 2008-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cocrystal structure guided array synthesis of PPARgamma inverse agonists
BIOORG.MED.CHEM.LETT., 17, 2007
|
|
2POC
 
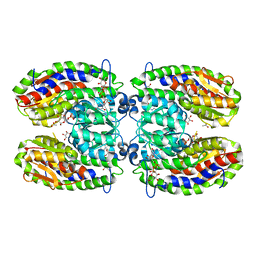 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETATE ION, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-04-26 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
2POD
 
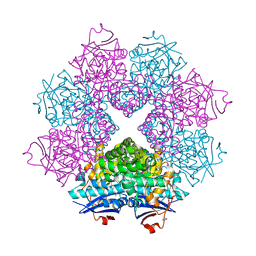 | | Crystal structure of a member of enolase superfamily from Burkholderia pseudomallei K96243 | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme, SODIUM ION | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Gilmore, J.M, Iizuka, M, Ozyurt, S, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-06-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of a Member of Enolase Superfamily from Burkholderia Pseudomallei.
To be Published
|
|
2POE
 
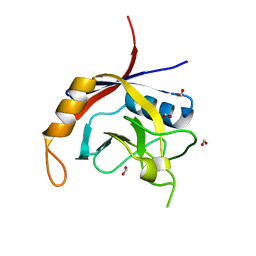 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 | | Descriptor: | Cyclophilin-like protein, putative, FORMIC ACID | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660.
To be Published
|
|
2POF
 
 | |
2POG
 
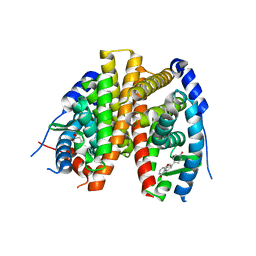 | | Benzopyrans as Selective Estrogen Receptor b Agonists (SERBAs). Part 2: Structure Activity Relationship Studies on the Benzopyran Scaffold. | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-9-OL, Estrogen receptor | | Authors: | Richardson, T.I, Norman, B.H, Lugar, C.W, Jones, S.A, Wang, Y, Durbin, J.D, Krishnan, V, Dodge, J.A. | | Deposit date: | 2007-04-26 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 2: structure-activity relationship studies on the benzopyran scaffold.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2POH
 
 | |
