6HM5
 
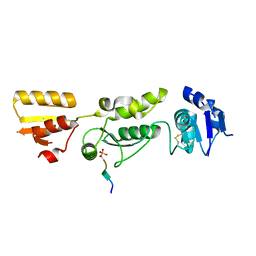 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a RAD9 phosphopeptide | | Descriptor: | Cell cycle checkpoint control protein RAD9A, DNA topoisomerase II binding protein 1 | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.330038 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
7PYH
 
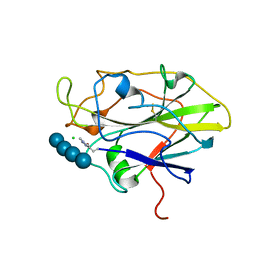 | | Structure of LPMO in complex with cellotetraose at 1.45x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
5JX2
 
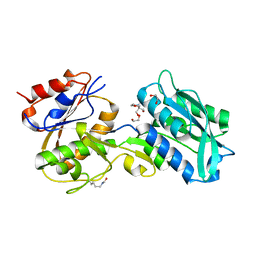 | | Crystal structure of MglB-2 (Tp0684) from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2016-05-12 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.0504 Å) | | Cite: | The Tp0684 (MglB-2) Lipoprotein of Treponema pallidum: A Glucose-Binding Protein with Divergent Topology.
Plos One, 11, 2016
|
|
6QA4
 
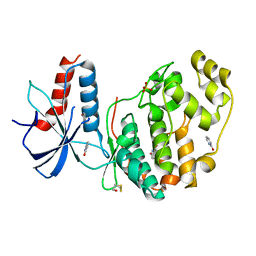 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-pyridin-2-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
8RZF
 
 | |
7AZY
 
 | |
8UG3
 
 | | Crystal structure of KHK-C and compound 23 | | Descriptor: | 2-[(4P)-4-{2-[(2S)-2-methylazetidin-1-yl]-6-(trifluoromethyl)pyrimidin-4-yl}-1H-pyrazol-1-yl]-1-(piperazin-1-yl)ethan-1-one, GLYCEROL, Ketohexokinase, ... | | Authors: | Durbin, J.D, Guo, S.Y. | | Deposit date: | 2023-10-05 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of LY3522348: A Highly Selective and Orally Efficacious Ketohexokinase Inhibitor.
J.Med.Chem., 66, 2023
|
|
4R7O
 
 | | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Crystal Structure of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus anthraci
To be Published
|
|
6Q2N
 
 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | Descriptor: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Y7J
 
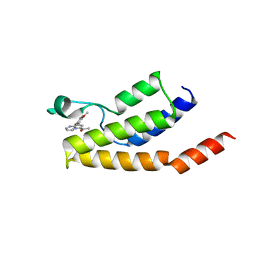 | | Structure of the BRD9 bromodomain and compound 15 | | Descriptor: | 1-[8-(2,5-dimethoxyphenyl)pyrrolo[1,2-a]pyrimidin-6-yl]ethanone, ACETATE ION, Bromodomain-containing protein 9, ... | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M. | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the BRD9 bromodomain
To Be Published
|
|
6Y7K
 
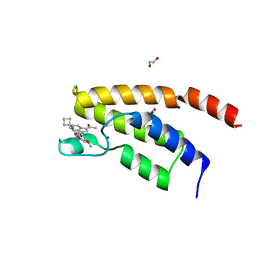 | | Structure of the BRD9 bromodomain and compound 27 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, SODIUM ION, ... | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M. | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the BRD9 bromodomain
To Be Published
|
|
8UD9
 
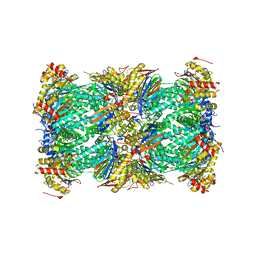 | | Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
9AY8
 
 | |
8PW1
 
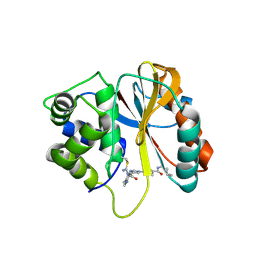 | | Structure of human UCHL1 in complex with CG341 inhibitor | | Descriptor: | (2~{S})-4-(iminomethyl)-1-methyl-~{N}-[1-[4-(pent-4-ynylcarbamoyl)phenyl]imidazol-4-yl]piperazine-2-carboxamide, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9HIR
 
 | | MnmG dimer within the MnmE-MnmG a4b2 complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme MnmG | | Authors: | Maes, L, Galicia, C, Fislage, M, Versees, W. | | Deposit date: | 2024-11-27 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-03 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Deciphering the RNA recognition by Musashi-1 to design protein and RNA variants for in vitro and in vivo applications.
Nucleic Acids Res., 53, 2025
|
|
7PYF
 
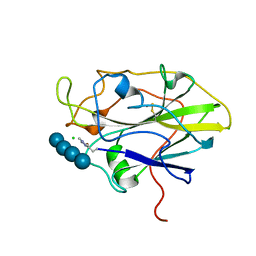 | | Structure of LPMO in complex with cellotetraose at 1.39x10^5 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6Q7G
 
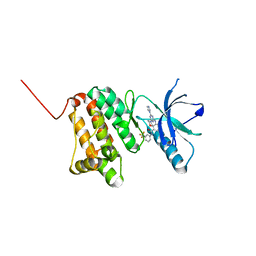 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATHA01 | | Descriptor: | 3-[(4-imidazol-1-yl-6-morpholin-4-yl-1,3,5-triazin-2-yl)amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.047 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
9GLY
 
 | |
8PTR
 
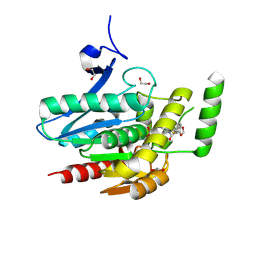 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5r | | Descriptor: | (3~{R},4~{S})-4-(1,3-benzodioxol-5-yl)-1-[1-(benzotriazol-1-ylcarbonyl)piperidin-4-yl]-3-(3-fluorophenyl)azetidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Butini, S, Benz, J, Grether, U, Leibrock, L, Papa, A, Maramai, S, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
8E3J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 4 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{6-[2-nitro-4-(pyrimidin-2-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7U6F
 
 | | Mouse retromer (VPS26/VPS35/VPS29) heterotrimers | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kendall, A.K, Chandra, M, Jackson, L.P. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Improved mammalian retromer cryo-EM structures reveal a new assembly interface.
J.Biol.Chem., 298, 2022
|
|
8E3P
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 5 | | Descriptor: | (2S,3S,4S,5R)-2-(hydroxymethyl)-1-{6-[3-nitro-5-(pyridin-4-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
4ZGH
 
 | | Structure of Sugar Binding Protein Pneumolysin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GOLD (I) CYANIDE ION, ... | | Authors: | Parker, M.W, Feil, S.C, Morton, C. | | Deposit date: | 2015-04-23 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae pneumolysin provides key insights into early steps of pore formation.
Sci Rep, 5, 2015
|
|
8ECW
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 11 | | Descriptor: | (2R,3R,4R,5S)-1-{2-[4-(2-{[(5M)-3-chloro-5-(pyridazin-3-yl)phenyl]amino}ethyl)phenyl]ethyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
5KF1
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, apo form, pH 5 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Holden, H.M, Thoden, J.B, Dopkins, B.J, tipton, P.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
