4R1G
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cloxacillin | | Descriptor: | CLOXACILLIN (OPEN FORM), Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cloxacillin
To be Published
|
|
4RA7
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-2-hydroxy-1-{[(2-hydroxynaphthalen-1-yl)carbonyl]amino}ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin
To be Published
|
|
4R3J
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin | | Descriptor: | (2R)-2-[(1R)-1-(acetylamino)-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin
To be Published
|
|
4R23
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin | | Descriptor: | (3R,4R,5R)-3-(2,6-dichlorophenyl)-N-{(1R)-1-[(2R,4S)-4-(dihydroxymethyl)-5,5-dimethyl-1,3-thiazolidin-2-yl]-2-oxoethyl} -5-methyl-1,2-oxazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-08 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with dicloxacillin
To be Published
|
|
3OC2
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCN
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCL
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with carbenicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3PBO
 
 | | Crystal structure of PBP3 complexed with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBT
 
 | | Crystal structure of PBP3 complexed with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 3 | | Authors: | Han, S, Evdokimov, A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBN
 
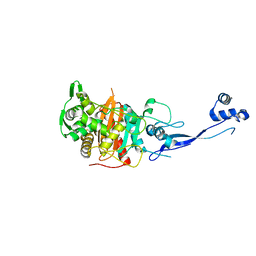 | | Crystal Structure of Apo PBP3 from Pseudomonas aeruginosa | | Descriptor: | Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBS
 
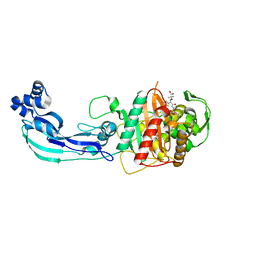 | | Crystal structure of PBP3 complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBQ
 
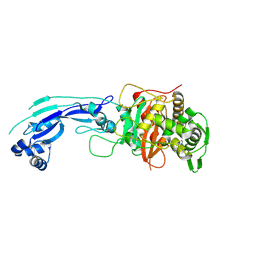 | | Crystal structure of PBP3 complexed with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBR
 
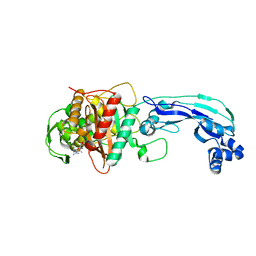 | | Crystal structure of PBP3 complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7ONY
 
 | | Crystal structure of PBP3 from P. aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONO
 
 | |
7ONZ
 
 | |
7ONW
 
 | | Crystal structure of PBP3 from E. coli in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONK
 
 | | Crystal structure of PBP3 from P. aeruginosa in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONN
 
 | | Crystal structure of PBP3 transpeptidase domain from E. coli in complex with AIC499 | | Descriptor: | (2S)-2-[(Z)-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2S)-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxy-3-[4-[N-[(3R)-piperidin-3-yl]carbamimidoyl]phenoxy]propanoic acid, DODECAETHYLENE GLYCOL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
7ONX
 
 | | Crystal structure of PBP3 from P. aeruginosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Freischem, S, Grimm, I, Weiergraeber, O.H. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Interaction Mode of the Novel Monobactam AIC499 Targeting Penicillin Binding Protein 3 of Gram-Negative Bacteria.
Biomolecules, 11, 2021
|
|
4WEJ
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted allyl monocarbam | | Descriptor: | (3R,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-[(5R)-5-hydroxy-4-oxo-4,5-dihydropyridin-2-yl]-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-3-(prop-2-en-1-yl)-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEL
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with SMC-3176 | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-3,10,10-trimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4WEK
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
1K25
 
 | |
