6JFJ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JFX
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltopentaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JEQ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with beta-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JHG
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121
To Be Published
|
|
6JHI
 
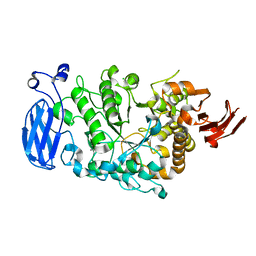 | | Crystal structure of mutant D470A of Pullulanase from Paenibacillus barengoltzii complexed with maltotetraose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pulullanase, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Crystal structure of mutant D470A of Pullulanase from Paenibacillus barengoltzii complexed with maltotetraose
To Be Published
|
|
6JHF
 
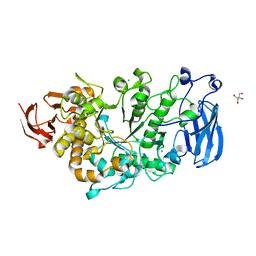 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JHH
 
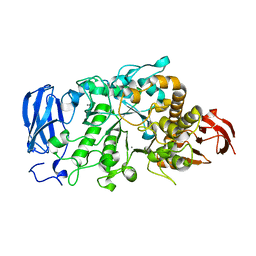 | | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose | | Descriptor: | CALCIUM ION, Pulullanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Crystal structure of mutant D350A of Pullulanase from Paenibacillus barengoltzii complexed with maltotriose
To Be Published
|
|
6FME
 
 | |
6IRS
 
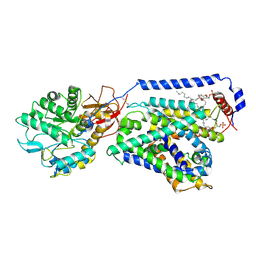 | | human LAT1-4F2hc complex incubated with JPH203 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Zhao, X, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex.
Nature, 568, 2019
|
|
6IRT
 
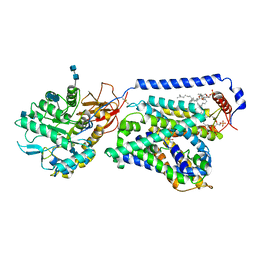 | | human LAT1-4F2hc complex bound with BCH | | Descriptor: | (1S,2S,4R)-2-aminobicyclo[2.2.1]heptane-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhao, X, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex.
Nature, 568, 2019
|
|
6I9Q
 
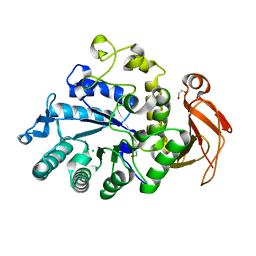 | | Structure of the mouse CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, CHLORIDE ION | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ectodomains of murine and human CD98hc.
Proteins, 87, 2019
|
|
6JMQ
 
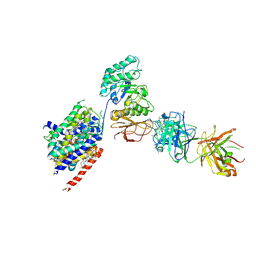 | | LAT1-CD98hc complex bound to MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Nakane, T, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JMR
 
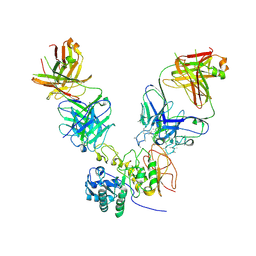 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6AAV
 
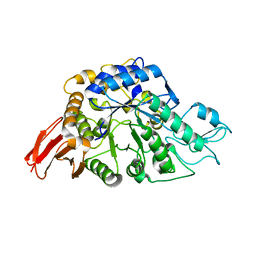 | | Crystal structure of alpha-glucosyl transfer enzyme, XgtA at 1.72 angstrom resolution | | Descriptor: | Alpha-glucosyltransferase | | Authors: | Kurumizaka, H, Arimura, Y, Kirimura, K, Watanabe, R. | | Deposit date: | 2018-07-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of alpha-glucosyl transfer enzyme XgtA from Xanthomonas campestris WU-9701.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
6S9V
 
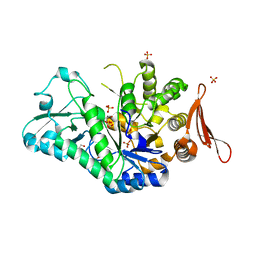 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Thermoanaerobacter thermosaccharolyticum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
6K5P
 
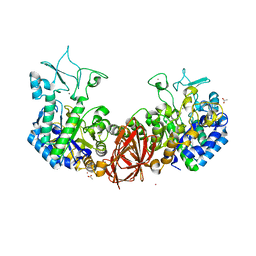 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
6L2H
 
 | | CGTase mutant-Y167H | | Descriptor: | Alpha-cyclodextrin glucanotransferase, CALCIUM ION | | Authors: | Fan, T.W, Hou, A.Q, Chao, Y.P, Sun, Y. | | Deposit date: | 2019-10-03 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structure basis of a mutant a-CGTase tyrosine167histidine from Bacillus sp. 602-1 with enhanced a-CD production
To Be Published
|
|
6SAV
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6IYG
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-15 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6J3X
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotriose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-01-06 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6OBX
 
 | |
6OCN
 
 | |
6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB | | Authors: | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | Deposit date: | 2019-03-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
6S8V
 
 | | Structure of the high affinity Anticalin P3D11 in complex with the human CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2019-07-10 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a high affinity Anticalin®directed against human CD98hc for theranostic applications.
Theranostics, 10, 2020
|
|
