5J6L
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with N-Butyl-5-[4-(2-fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | Heat shock protein HSP 90-alpha, N-butyl-5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methylbenzamide | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
6XMM
 
 | | Human aldolase A I98S | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, PHOSPHATE ION | | Authors: | Meneely, K.M, Brewer, K, Lamb, A.L. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substitutions at a rheostat position in human aldolase A cause a shift in the conformational population.
Protein Sci., 31, 2022
|
|
7U89
 
 | | Product of 14mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
3H8S
 
 | |
6XML
 
 | | Human aldolase A I98C | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, PHOSPHATE ION | | Authors: | Meneely, K.M, Brewer, K, Lamb, A.L. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substitutions at a rheostat position in human aldolase A cause a shift in the conformational population.
Protein Sci., 31, 2022
|
|
8YW6
 
 | | Cryo-EM structure of apo human mitochondrial pyruvate carrier in the IMS-open conformation at pH 8.0 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-29 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
6GFZ
 
 | | pVHL:EloB:EloC in complex with modified VH032 containing (3S,4S)-3-fluoro-4-hydroxyproline (ligand 14b) | | Descriptor: | (2~{R},3~{S},4~{S})-1-[(2~{S})-2-acetamido-3,3-dimethyl-butanoyl]-3-fluoranyl-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
6PDP
 
 | | Human PIM1 bound to benzothiophene inhibitor 379 | | Descriptor: | 5-[2-(acetylamino)-1-benzothiophen-4-yl]-N-cyclopropylthiophene-2-carboxamide, Peptide, SULFATE ION, ... | | Authors: | Godoi, P.H.C, Sriranganadane, D, Santiago, A.S, Fala, A.M, Ramos, P.Z, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human PIM1
To Be Published
|
|
8OFA
 
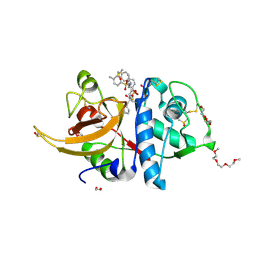 | | Crystal structure of human cathepsin L interacting with tosyl phenylalanyl chloromethyl ketone (TPCK) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 4-methyl-~{N}-[(2~{S})-4-oxidanyl-3-oxidanylidene-1-phenyl-butan-2-yl]benzenesulfonamide, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Ewert, W, Reinke, P.Y.A, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-03-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8I83
 
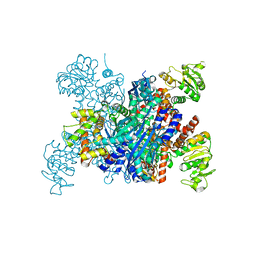 | |
6GI6
 
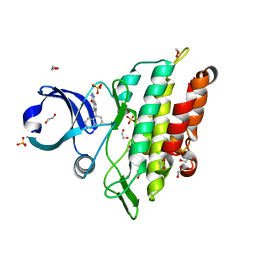 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
9DM6
 
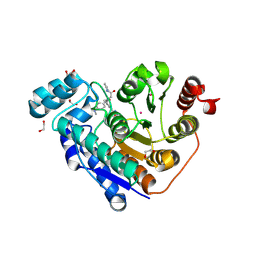 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with an ethylhydrazide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)-N-{[4-(2-ethylhydrazine-1-carbonyl)phenyl]methyl}benzamide, ... | | Authors: | Gaynes, M.N, Watson, P.R, Konig, B, Christianson, D.W. | | Deposit date: | 2024-09-12 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploring Alternative Zinc-Binding Groups in Histone Deacetylase (HDAC) Inhibitors Uncovers DS-103 as a Potent Ethylhydrazide-Based HDAC Inhibitor with Chemosensitizing Properties.
J.Med.Chem., 68, 2025
|
|
8R7E
 
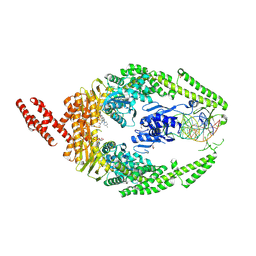 | | MutSbeta bound to compound CHDI-00898647 in the canonical DNA-mismatch bound form | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Abivertinib, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Steinbacher, S, Brace, G, Tillack, K, Johnson, P, Ritzefeld, M, Schaertl, S, Frush, E, Warfield, B, Ballantyne, G, Lee, J.-H, Witte, D, Finley, M, Prasad, B, Monteagudo, E, Plotnikov, N, Pacifici, R, Maillard, M, Wilkinson, H, Iyer, R, Dominguez, C, Vogt, T, Felsenfeld, D, Haque, T. | | Deposit date: | 2023-11-24 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Identification of orthosteric inhibitors of MutSbeta ATPase function
Chemrxiv, 2024
|
|
8FV2
 
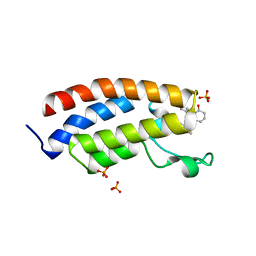 | | Bromodomain of CBP liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
6GND
 
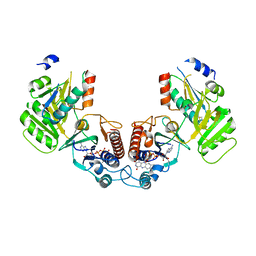 | | Crystal structure of the complex of a Ferredoxin-Flavin Thioredoxin Reductase and a Thioredoxin from Clostridium acetobutylicum at 2.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Balsera, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-12-12 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6SWX
 
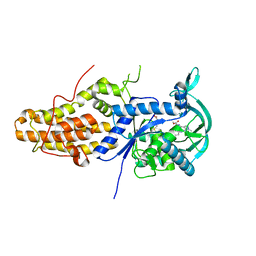 | | Leishmania major methionyl-tRNA synthetase in complex with an allosteric inhibitor | | Descriptor: | METHIONINE, Putative methionyl-tRNA synthetase, methyl 2-[[6-[[3,4-bis(fluoranyl)phenyl]amino]-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl]amino]ethanoate | | Authors: | Robinson, D.A, Torrie, L.S, Shepherd, S.M, De Rycker, M, Thomas, M.G, Wyatt, P.G. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of an Allosteric Binding Site in Kinetoplastid Methionyl-tRNA Synthetase.
Acs Infect Dis., 6, 2020
|
|
8QZ7
 
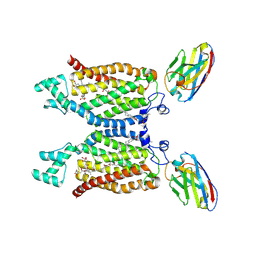 | | Structure of human ceramide synthase 6 (CerS6) in complex with N-palmitoyl fumonisin B1 | | Descriptor: | (2~{R})-2-[2-[(5~{R},6~{R},7~{S},9~{S},11~{R},16~{R},18~{S},19~{S})-6-[(3~{R})-3-carboxy-5-oxidanyl-5-oxidanylidene-pentanoyl]oxy-19-(hexadecanoylamino)-5,9-dimethyl-11,16,18-tris(oxidanyl)icosan-7-yl]oxy-2-oxidanylidene-ethyl]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4XYQ
 
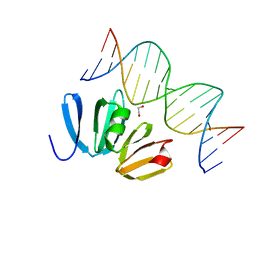 | | Structure of AgrA LytTR domain in complex with promoters | | Descriptor: | 1,2-ETHANEDIOL, Accessory gene regulator A, DNA (5'-D(*AP*AP*TP*AP*CP*TP*TP*AP*AP*CP*TP*GP*TP*TP*AP*A)-3'), ... | | Authors: | Gopal, B, Rajasree, K. | | Deposit date: | 2015-02-03 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational features of theStaphylococcus aureusAgrA-promoter interactions rationalize quorum-sensing triggered gene expression.
Biochem Biophys Rep, 6, 2016
|
|
6N2K
 
 | | Tetrahydropyridopyrimidines as Covalent Inhibitors of KRAS-G12C | | Descriptor: | 1-{4-[2-{[(2R)-1-(dimethylamino)propan-2-yl]oxy}-7-(3-hydroxynaphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P. | | Deposit date: | 2018-11-13 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity.
ACS Med Chem Lett, 9, 2018
|
|
5JF2
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT002 | | Descriptor: | (3R)-3-{3-[(4-fluorophenyl)methyl]-1,2,4-oxadiazol-5-yl}-N-hydroxyheptanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
7R2I
 
 | |
9JXE
 
 | | Cryo-EM structure of apo human XPR1, class 1, with one visible SPX domain | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Wang, X, Bai, Z, Wallis, C, Wang, H, Han, Y, Jin, R, Lei, M, Gu, C, Jessen, H, Shears, S, Sun, Y, Corry, B, Zhang, Y. | | Deposit date: | 2024-10-11 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | KIDINS220 and InsP8 safeguard the stepwise regulation of phosphate exporter XPR1.
Mol.Cell, 85, 2025
|
|
6SZH
 
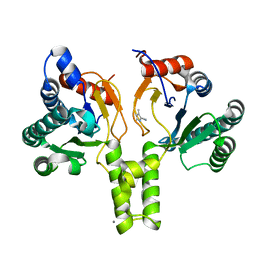 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GW197 | | Descriptor: | 3,5-dimethyl-1~{H}-pyrrole-2-carbonitrile, CALCIUM ION, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8FUP
 
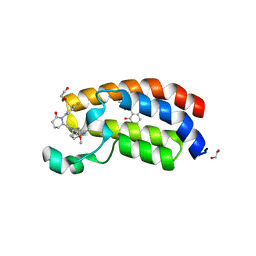 | | Bromodomain of CBP liganded with BMS-536924 and CCS-1477 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
8AYX
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
