6AMW
 
 | |
6AMV
 
 | |
4WP4
 
 | | Hev b 6.02 (hevein) extracted from surgical gloves | | Descriptor: | Pro-hevein, THIOUREA | | Authors: | Galicia, C, Rodriguez-Romero, A. | | Deposit date: | 2014-10-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Impact of the vulcanization process on the structural characteristics and IgE recognition of two allergens, Hev b 2 and Hev b 6.02, extracted from latex surgical gloves.
Mol.Immunol., 65, 2015
|
|
8B30
 
 | |
8ADQ
 
 | | Crystal structure of holo-SwHPA-Mg (hydroxy ketone aldolase) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate and D-Glyceraldehyde | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, D-Glyceraldehyde, ... | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8BGN
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Diacetylchitobiose deacetylase, ... | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8BGP
 
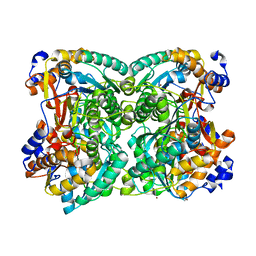 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus anomalous data | | Descriptor: | Diacetylchitobiose deacetylase, ZINC ION | | Authors: | Rypniewski, W, Biniek-Antosiak, K, Bejger, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8BGO
 
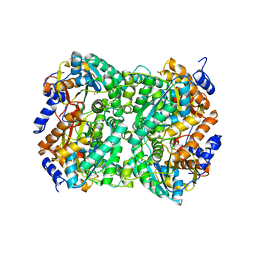 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus with substrate N,N-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Diacetylchitobiose deacetylase, ZINC ION | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
2KM3
 
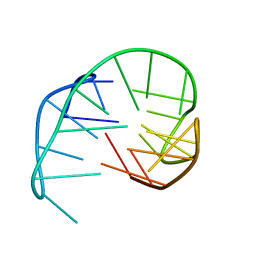 | | Structure of an intramolecular G-quadruplex containing a G.C.G.C tetrad formed by human telomeric variant CTAGGG repeats | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*TP*AP*GP*GP*GP*CP*TP*AP*GP*GP*GP*CP*TP*AP*GP*GP*G)-3') | | Authors: | Lim, K.W, Alberti, P, Guedin, A, Lacroix, L, Riou, J.F, Royle, N.J, Mergny, J.L, Phan, A.T. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad
Nucleic Acids Res., 37, 2009
|
|
2HVZ
 
 | | Solution structure of the RRM domain of SR rich factor 9G8 | | Descriptor: | Splicing factor, arginine/serine-rich 7 | | Authors: | Tintaru, A.M, Hautbergue, G.M, Golovanov, A.P, Lian, L.Y, Wilson, S.A. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.
Embo J., 25, 2006
|
|
2JQR
 
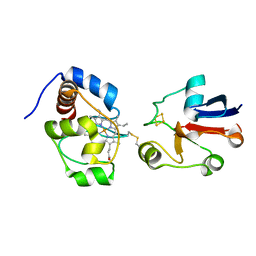 | | Solution model of crosslinked complex of cytochrome c and adrenodoxin | | Descriptor: | Adrenodoxin, mitochondrial, Cytochrome c iso-1, ... | | Authors: | Xu, X, Reinle, W, Hannemann, F, Konarev, P.V, Svergun, D.I, Bernhardt, R, Ubbink, M. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy
J.Am.Chem.Soc., 130, 2008
|
|
2JWO
 
 | | A PHD finger motif in the C-terminus of RAG2 modulates recombination activity | | Descriptor: | V(D)J recombination-activating protein 2, ZINC ION | | Authors: | Ivanov, D, Hyberts, S.G, Sun, Z, Wagner, G. | | Deposit date: | 2007-10-17 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A PHD finger motif in the C terminus of RAG2 modulates recombination activity.
J.Biol.Chem., 280, 2005
|
|
2LRE
 
 | | The solution structure of the dimeric Acanthaporin | | Descriptor: | Acanthaporin | | Authors: | Michalek, M, Soennichsen, F.D, Wechselberger, R, Dingley, A.J, Wienk, H, Simanski, M, Herbst, R, Lorenzen, I, Marciano-Cabral, F, Gelhaus, C, Groetzinger, J, Leippe, M. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Nat.Chem.Biol., 9, 2013
|
|
2LJL
 
 | |
4KII
 
 | | Beta-lactoglobulin in complex with Cp*Rh(III)Cl N,N-di(pyridin-2-yl)dodecanamide | | Descriptor: | Beta-lactoglobulin, [N,N-di(pyridin-2-yl-kappaN)dodecanamide]rhodium | | Authors: | Cherrier, M.V, Amara, P, Engilberge, S, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-02 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Enantioselectivity in the Transfer Hydrogenation of a Ketone Catalyzed by an Artificial Metalloenzyme
Eur.J.Inorg.Chem., 21, 2013
|
|
6RLZ
 
 | |
4X8H
 
 | | Crystal structure of E. coli Adenylate kinase P177A mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
4X8L
 
 | | Crystal structure of E. coli Adenylate kinase P177A mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
1N3K
 
 | | Solution structure of phosphoprotein enriched in astrocytes 15 kDa (PEA-15) | | Descriptor: | Astrocytic phosphoprotein PEA-15 | | Authors: | Hill, J.M, Vaidyanathan, H, Ramos, J.W, Ginsberg, M.H, Werner, M.H. | | Deposit date: | 2002-10-28 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of ERK MAP Kinase by PEA-15 Reveals a Common Docking Site Within the Death Domain and Death Effector Domain
Embo J., 21, 2002
|
|
7TIO
 
 | |
1KO9
 
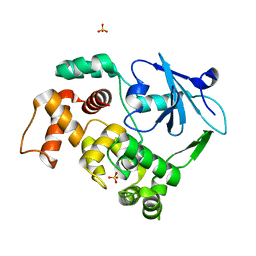 | | Native Structure of the Human 8-oxoguanine DNA Glycosylase hOGG1 | | Descriptor: | 8-oxoguanine DNA glycosylase, SULFATE ION | | Authors: | Bjoras, M, Seeberg, E, Luna, L, Pearl, L.H, Barrett, T.E. | | Deposit date: | 2001-12-20 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reciprocal "flipping" underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase.
J.Mol.Biol., 317, 2002
|
|
7TIR
 
 | |
7TIS
 
 | |
7TIP
 
 | |
7TIQ
 
 | |
