5L98
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 3-amino-2-methylpyridine derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[(1~{S})-1-(2,3-dihydro-1,4-benzodioxin-6-yl)ethyl]-2-methyl-pyridin-3-amine | | Authors: | Lolli, G, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2016-06-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Derivatives of 3-Amino-2-methylpyridine as BAZ2B Bromodomain Ligands: In Silico Discovery and in Crystallo Validation.
J. Med. Chem., 59, 2016
|
|
8STW
 
 | | K384N HUMAN CYSTATHIONINE BETA-SYNTHASE (delta 411-551) | | Descriptor: | Cystathionine beta-synthase, K384N variant, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mascarenhas, R, Roman, J, Banerjee, R. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Disease-causing cystathionine beta-synthase linker mutations impair allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8SM3
 
 | | Structure of Bacillus cereus VD045 Gabija GajA-GajB Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB, SULFATE ION | | Authors: | Antine, S.P, Mooney, S.E, Johnson, A.G, Kranzusch, P.J. | | Deposit date: | 2023-04-25 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
7CC8
 
 | | Crystal structure of White Spot Syndrome Virus Thymidylate Synthase - Apo form | | Descriptor: | SULFATE ION, TRIETHYLENE GLYCOL, Thymidylate Synthase | | Authors: | Kumar, S, Panchal, N.V, Shaikh, N, Vasudevan, D. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements.
Int.J.Biol.Macromol., 167, 2021
|
|
8SXS
 
 | |
5LBK
 
 | |
4U3G
 
 | | Crystal structure of Escherichia coli bacterioferritin mutant D132F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Wong, S.G, Grigg, J.C, Le Brun, N.E, Moore, G.R, Murphy, M.E.P, Mauk, A.G. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The B-type Channel Is a Major Route for Iron Entry into the Ferroxidase Center and Central Cavity of Bacterioferritin.
J.Biol.Chem., 290, 2015
|
|
8SMF
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-3' gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Gp34.65, MAGNESIUM ION | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
4U3O
 
 | | Octameric RNA duplex soaked in manganese(II)chloride | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
5KSO
 
 | | hMiro1 C-domain GDP-Pi Complex P3121 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1, PHOSPHATE ION | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
4UC3
 
 | | Translocator protein 18 kDa (TSPO) from Rhodobacter sphaeroides wild type | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
8SMH
 
 | |
5KTI
 
 | | Structure of cow mincle complexed with KMJ1 | | Descriptor: | 2,3-dimethoxybenzoic acid, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Feinberg, H, Rambaruth, N.D.S, Jegouzo, S.A.F, Jacobsen, K.M, Djurhuus, R, Poulsen, T.B, Taylor, M.E, Drickamer, K, Weis, W.I. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle.
J.Biol.Chem., 291, 2016
|
|
8SP1
 
 | |
8SMJ
 
 | |
5KTV
 
 | |
8STS
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STV
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) variant in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600), a non-nucleoside inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, MAGNESIUM ION, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8SME
 
 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
5L8I
 
 | | crystal structure of human FABP6 apo-protein | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, Gastrotropin, ... | | Authors: | Hendrick, A, Mueller, I, Leonard, P.M, Davenport, R, Mitchell, P. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification and Investigation of Novel Binding Fragments in the Fatty Acid Binding Protein 6 (FABP6).
J.Med.Chem., 59, 2016
|
|
5KUO
 
 | | Human cyclophilin A at 100K, Data set 3 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
7CU2
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES ALBOGRISEOLUS FLAVIN-DEPENDENT TRYPTOPHAN 6-HALOGENASE THAL IN COMPLEX WITH REDUCED FAD | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
8T1U
 
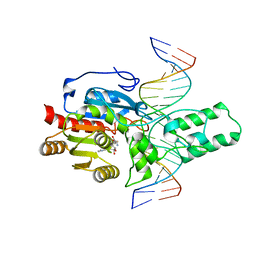 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
5KUV
 
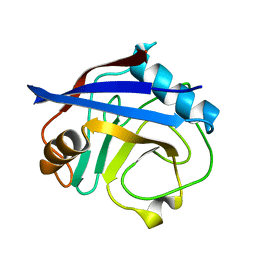 | | Human cyclophilin A at 100K, Data set 8 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5L99
 
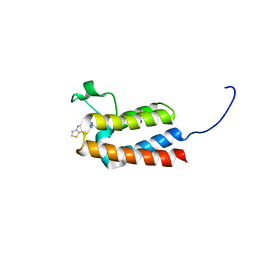 | |
