4IAI
 
 | |
4IB3
 
 | |
2AYU
 
 | |
4IB1
 
 | |
4IAD
 
 | |
4IAZ
 
 | |
4IB0
 
 | |
7QJ9
 
 | |
7QJ6
 
 | |
8DGS
 
 | | Cryo-EM structure of a RAS/RAF complex (state 1) | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Eck, M.J, Jeon, H, Park, E, Rawson, S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of a RAS/RAF recruitment complex.
Nat Commun, 14, 2023
|
|
8DGT
 
 | | Cryo-EM structure of a RAS/RAF complex (state 2) | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Eck, M.J, Jeon, H, Park, E, Rawson, S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a RAS/RAF recruitment complex.
Nat Commun, 14, 2023
|
|
8YD8
 
 | |
8YD7
 
 | | Structure of FADD/Caspase-8/cFLIP death effector domain assembly | | Descriptor: | CASP8 and FADD-like apoptosis regulator subunit p12, Caspase-8, FAS-associated death domain protein, ... | | Authors: | Lin, S.-C, Yang, C.-Y. | | Deposit date: | 2024-02-19 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Deciphering DED assembly mechanisms in FADD-procaspase-8-cFLIP complexes regulating apoptosis.
Nat Commun, 15, 2024
|
|
8YBX
 
 | |
4AYA
 
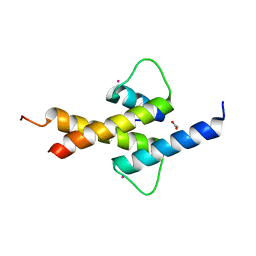 | | Crystal structure of ID2 HLH homodimer at 2.1A resolution | | Descriptor: | ACETATE ION, DNA-BINDING PROTEIN INHIBITOR ID-2, POTASSIUM ION | | Authors: | Wong, M.V, Jiang, S, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-06-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Divalent Ion is Crucial in the Structure and Dominant-Negative Function of Id Proteins, a Class of Helix-Loop-Helix Transcription Regulators.
Plos One, 7, 2012
|
|
4MD9
 
 | |
8P81
 
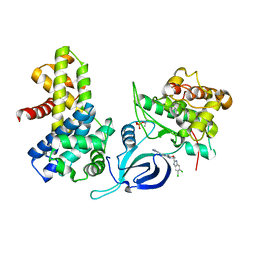 | | Crystal structure of human Cdk12/Cyclin K in complex with inhibitor SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, ~{N}-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(1-methylpyrazol-4-yl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2023-05-31 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The reversible inhibitor SR-4835 binds Cdk12/cyclin K in a noncanonical G-loop conformation.
J.Biol.Chem., 300, 2023
|
|
2MEZ
 
 | | Flexible anchoring of archaeal MBF1 on ribosomes suggests role as recruitment factor | | Descriptor: | Multiprotein Bridging Factor (MBP-like) | | Authors: | Launay, H, Blombarch, F, Camilloni, C, Vendruscolo, M, van des Oost, J, Christodoulou, J. | | Deposit date: | 2013-10-03 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain.
Biochem.J., 462, 2014
|
|
5ZHX
 
 | |
4IL1
 
 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
5UQ3
 
 | |
5BQD
 
 | | Crystal Structure of TBX5 (1-239) Dimer | | Descriptor: | MAGNESIUM ION, T-box transcription factor TBX5 | | Authors: | Pradhan, L, Gopal, S, Patel, A, Kasahara, H, Nam, H.J. | | Deposit date: | 2015-05-28 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.
Biochemistry, 55, 2016
|
|
4IAK
 
 | |
4IAY
 
 | |
1PFL
 
 | | REFINED SOLUTION STRUCTURE OF HUMAN PROFILIN I | | Descriptor: | PROFILIN I | | Authors: | Metzler, W.J, Farmer II, B.T, Constantine, K.L, Friedrichs, M.S, Lavoie, T, Mueller, L. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of human profilin I.
Protein Sci., 4, 1995
|
|
