5PZL
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 2-({3-[1-(2-CYCLOPROPYLETHYL)-6-FLUORO-4-HYDROXY-2-OXO-1,2-DIHYDROQUINOLIN-3-YL]-1,1-DIOXO-1,4-DIHYDRO-1LAMBDA~6~,2,4-BENZOTHIADIAZIN-7-YL}OXY)ACETAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-({3-[1-(2-cyclopropylethyl)-6-fluoro-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxo-1,4-dihydro-1lambda~6~,2,4-benzothiadiazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
5PZK
 
 | |
5PZP
 
 | |
5PTD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT H32A | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
6LBN
 
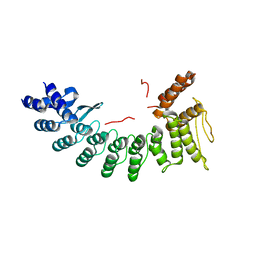 | | Structure of SIL1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEN
 
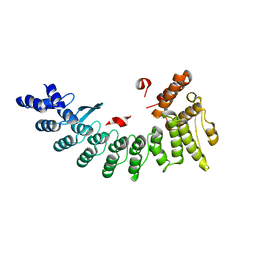 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEY
 
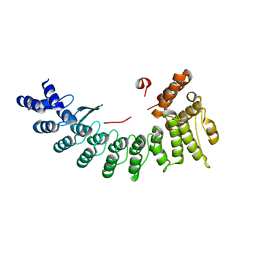 | | Structure of Sil1G bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Nucleotide exchange factor SIL1 | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LF0
 
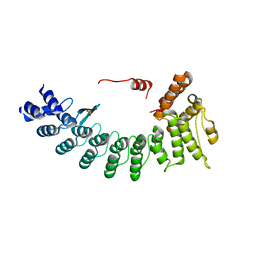 | | Structure of FEM1C | | Descriptor: | Protein fem-1 homolog C, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
9N9S
 
 | | Model of APC/C-CDC20-UBE2C from H3/H4-bound complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Skrajna, A, Bodrug, T, Brown, N.G, McGinty, R.K. | | Deposit date: | 2025-02-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | APC/C-mediated ubiquitylation of extranucleosomal histone complexes lacking canonical degrons.
Nat Commun, 16, 2025
|
|
9N9R
 
 | | Model of APC/C-CDC20-UBE2C from H2A/H2B-bound complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Skrajna, A, Bodrug, T, Brown, N.G, McGinty, R.K. | | Deposit date: | 2025-02-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | APC/C-mediated ubiquitylation of extranucleosomal histone complexes lacking canonical degrons.
Nat Commun, 16, 2025
|
|
6LDP
 
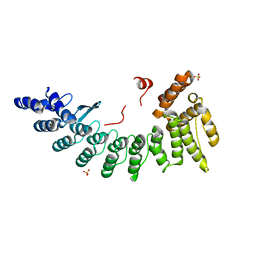 | | Structure of CDK5R1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
7PTD
 
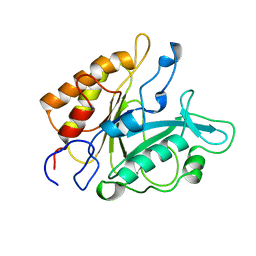 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT R163K | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
7NZY
 
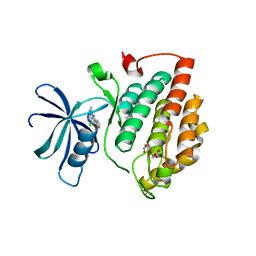 | | Crystal structure of human Casein Kinase I delta in complex with CGS-15943 | | Descriptor: | 9-chloranyl-2-(furan-2-yl)-[1,2,4]triazolo[1,5-c]quinazolin-5-amine, Casein kinase I isoform delta, MALONATE ION, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2021-03-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phenotypic Discovery of Triazolo[1,5- c ]quinazolines as a First-In-Class Bone Morphogenetic Protein Amplifier Chemotype.
J.Med.Chem., 65, 2022
|
|
6ZJG
 
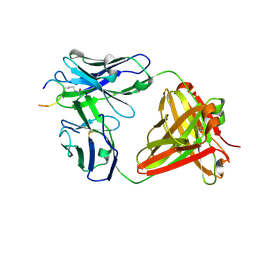 | | Crystal structure of ACPA E4 in complex with CII-C-48-CIT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACPA E4 Fab fragment - heavy chain, ACPA E4 Fab fragment - light chain, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of ACPA E4 in complex with CII-C-48-CIT
To Be Published
|
|
8VJ1
 
 | | Structure of C-terminal domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
7ANQ
 
 | | Complete PCSK9 C-ter domain in complex with VHH P1.40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Proprotein convertase subtilisin/kexin type 9, SULFATE ION, ... | | Authors: | Ciccone, L, Legrand, P, Stura, E.A, Dive, V, Seidahn, N.G, Fruchart Gaillard, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular interactions of PCSK9 with an inhibitory nanobody, CAP1 and HLA-C: Functional regulation of LDLR levels.
Mol Metab, 67, 2022
|
|
8CK8
 
 | |
8CK3
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (S)-1-(3,5-Difluoro-phenyl)-5,5-difluoro-3-methanesulfonyl-5,6-dihydro-4H-cyclopenta[c]thiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-4,6-dihydrocyclopenta[c]thiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Lehmannn, M, Diehl, L. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
8F8N
 
 | |
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
6FM6
 
 | | Crystal structure of the class C beta-lactamase TRU-1 from Aeromonas enteropelogenes | | Descriptor: | Beta-lactamase, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Pozzi, C, De Luca, F, Di Pisa, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic-Resolution Structure of a Class C beta-Lactamase and Its Complex with Avibactam.
ChemMedChem, 13, 2018
|
|
6FM7
 
 | | Crystal structure of the class C beta-lactamase TRU-1 from Aeromonas enteropelogenes in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Di Pisa, F, Docquier, J.D, Mangani, S. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic-Resolution Structure of a Class C beta-Lactamase and Its Complex with Avibactam.
ChemMedChem, 13, 2018
|
|
6LUS
 
 | | Crystal structure of the Mengla Virus VP30 C-terminal domain | | Descriptor: | Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Wen, K.N, Chu, H.G, Wang, C.H, Qin, X.C. | | Deposit date: | 2020-01-30 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Mengla virus VP30 C-terminal domain.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6GD7
 
 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with PEG | | Descriptor: | Cytochrome c iso-1, GLYCEROL, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8EDE
 
 | | Crystal structure of covalent inhibitor 2-chloro-N'-(N-(4-chlorophenyl)-N-methylglycyl)acetohydrazide bound to Ubiquitin C-terminal Hydrolase-L1 | | Descriptor: | 2-[(4-chlorophenyl)-methyl-amino]-~{N}'-ethanoyl-ethanehydrazide, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Patel, R, Imhoff, R, Flaherty, D, Das, C. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Covalent Fragment Screening and Optimization Identifies the Chloroacetohydrazide Scaffold as Inhibitors for Ubiquitin C-terminal Hydrolase L1.
J.Med.Chem., 67, 2024
|
|
