1WNZ
 
 | |
4K88
 
 | | Crystal structure of human prolyl-tRNA synthetase (halofuginone bound form) | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Proline--tRNA ligase, ZINC ION | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K87
 
 | | Crystal structure of human prolyl-tRNA synthetase (substrate bound form) | | Descriptor: | ADENOSINE, PROLINE, Proline--tRNA ligase, ... | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1XTY
 
 | | Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Fromant, M, Schmitt, E, Mechulam, Y, Lazennec, C, Plateau, P, Blanquet, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and identification of active site residues of Sulfolobus solfataricus peptidyl-tRNA hydrolase.
Biochemistry, 44, 2005
|
|
4KQE
 
 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
1TJE
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase | | Descriptor: | Threonyl-tRNA synthetase | | Authors: | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
1SRY
 
 | |
2ZIM
 
 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Steitz, T.A, Kavran, J.M. | | Deposit date: | 2008-02-19 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ZZG
 
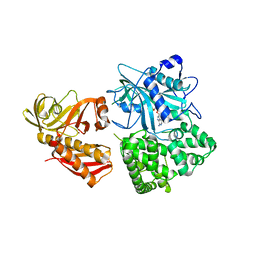 | | Crystal structure of alanyl-tRNA synthetase in complex with 5''-O-(N-(L-alanyl)-sulfamyoxyl) adenine without oligomerization domain | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1QE0
 
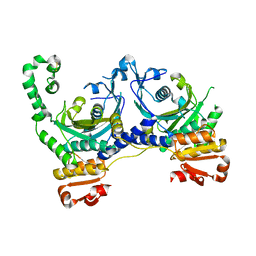 | | CRYSTAL STRUCTURE OF APO S. AUREUS HISTIDYL-TRNA SYNTHETASE | | Descriptor: | Histidine--tRNA ligase | | Authors: | Qiu, X, Janson, C.A, Blackburn, M.N, Chohan, I.K, Hibbs, M, Abdel-Meguid, S.S. | | Deposit date: | 1999-07-12 | | Release date: | 2000-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative structural dynamics and a novel fidelity mechanism in histidyl-tRNA synthetases.
Biochemistry, 38, 1999
|
|
4K47
 
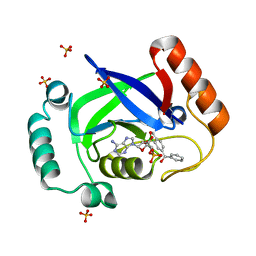 | | Structure of the Streptococcus pneumoniae leucyl-tRNA synthetase editing domain bound to a benzoxaborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1R,5R,6R,8S)-6-(6-aminopurin-9-yl)-3'-[(R)-oxidanyl(phenyl)methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1,3,5-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Hu, Q.H, Liu, R.J, Fang, Z.P, Zhang, J, Ding, Y.Y, Tan, M, Wang, M, Pan, W, Zhou, H.C, Wang, E.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase
Sci Rep, 3, 2013
|
|
4K48
 
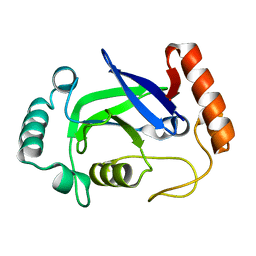 | | Structure of the Streptococcus pneumoniae leucyl-tRNA synthetase editing domain | | Descriptor: | Leucine--tRNA ligase | | Authors: | Hu, Q.H, Liu, R.J, Fang, Z.P, Zhang, J, Ding, Y.Y, Tan, M, Wang, M, Pan, W, Zhou, H.C, Wang, E.D. | | Deposit date: | 2013-04-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase
Sci Rep, 3, 2013
|
|
2JVA
 
 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | Descriptor: | Peptidyl-tRNA hydrolase domain protein | | Authors: | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
2ZIO
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AlocLys-AMP and PNP | | Descriptor: | 5'-O-[(S)-({(2S)-2-amino-6-[(propoxycarbonyl)amino]hexanoyl}oxy)(hydroxy)phosphoryl]adenosine, IMIDODIPHOSPHORIC ACID, Pyrrolysyl-tRNA synthetase | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-19 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
2ZIN
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and an ATP analogue | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-19 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
1QZA
 
 | | Coordinates of the A/T site tRNA model fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZB
 
 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
2X1B
 
 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2O33
 
 | | Solution structure of U2 snRNA stem I from S. cerevisiae | | Descriptor: | U2 snRNA | | Authors: | Sashital, D.G, Venditti, V, Angers, C.A, Cornilescu, G, Butcher, S.E. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human.
Rna, 13, 2007
|
|
3JA1
 
 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
1U1Y
 
 | | Crystal structure of a complex between WT bacteriophage MS2 coat protein and an F5 aptamer RNA stemloop with 2aminopurine substituted at the-10 position | | Descriptor: | 5'-R(*CP*CP*GP*GP*(2PR)P*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*G)-3', Coat protein | | Authors: | Horn, W.T, Convery, M.A, Stonehouse, N.J, Adams, C.J, Liljas, L, Phillips, S.E, Stockley, P.G. | | Deposit date: | 2004-07-16 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand-RNA interactions.
Rna, 10, 2004
|
|
4CS3
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in complex with an adenylated furan-bearing noncanonical amino acid and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(furan-2-yl)ethyl hydrogen carbonate, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
9MTT
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with mRNA, A-site Q230-N5-methylated Release Factor 1, and P-site deacylated-tRNAcys at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Basu, R.S, Vassilevski, A.A, Gagnon, M.G, Polikanov, Y.S. | | Deposit date: | 2025-01-12 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of release factor-mediated peptidyl-tRNA hydrolysis on the ribosome.
Science, 388, 2025
|
|
5LZS
 
 | | Structure of the mammalian ribosomal elongation complex with aminoacyl-tRNA, eEF1A, and didemnin B | | Descriptor: | (2~{S})-~{N}-[(2~{R})-1-[[(3~{S},6~{S},8~{S},12~{S},13~{R},16~{S},17~{R},20~{S},23~{S})-13-[(2~{S})-butan-2-yl]-20-[(4-methoxyphenyl)methyl]-6,17,21-trimethyl-3-(2-methylpropyl)-12-oxidanyl-2,5,7,10,15,19,22-heptakis(oxidanylidene)-8-propan-2-yl-9,18-dioxa-1,4,14,21-tetrazabicyclo[21.3.0]hexacosan-16-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-~{N}-methyl-1-[(2~{S})-2-oxidanylpropanoyl]pyrrolidine-2-carboxamide, 18S ribosomal RNA, 28S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5XIO
 
 | | Crystal Structure of Cryptosporidium parvum Prolyl-tRNA Synthetase (CpPRS) in complex with Halofuginone | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Proline-tRNA synthetase class II aaRS (Ybak RNA binding domain plus tRNA synthetase), ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
