6P14
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form B) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, ACETATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93001127 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6PDY
 
 | | Msp1-substrate complex in open conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6P13
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form A) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6PEK
 
 | | Structure of Spastin Hexamer (Subunit A-E) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6PDW
 
 | | Msp1-substrate complex in closed conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PEN
 
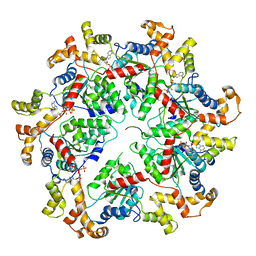 | | Structure of Spastin Hexamer (whole model) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, EYEYEYEYEY, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6PXI
 
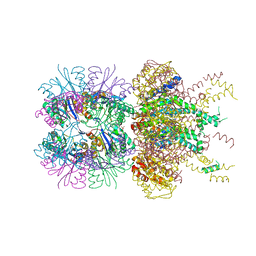 | | The crystal structure of a singly capped HslUV complex with an axial pore plug and a HslU E257Q mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.447 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6PXL
 
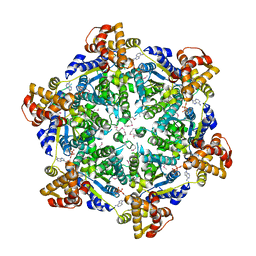 | | 3.74 Angstroms resolution structure of HlsU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, MAGNESIUM ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.741 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6PXK
 
 | | 3.65 Angstroms resolution structure of HslU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, SULFATE ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.647 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6QS8
 
 | |
6OG1
 
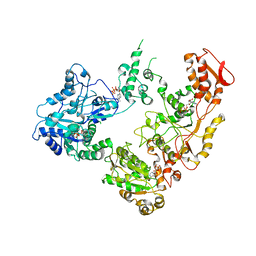 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6QS4
 
 | |
6OAB
 
 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6QS7
 
 | |
6OPC
 
 | | Cdc48 Hexamer in a complex with substrate and Shp1(Ubx Domain) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
6QS6
 
 | |
7Y53
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Z6H
 
 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-05-04 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
7YKT
 
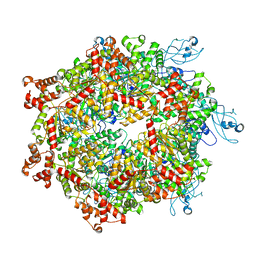 | | Cryo-EM structure of Drg1 hexamer in helical state treated with ADP/AMPPNP/benzo-diazaborine | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKZ
 
 | | Cryo-EM structure of Drg1 hexamer in the planar state treated with ADP/AMPPNP/Diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKL
 
 | | Cryo-EM structure of Drg1 hexamer treated with AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YKK
 
 | | Cryo-EM structure of Drg1 hexamer treated with ADP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2022-07-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7YPJ
 
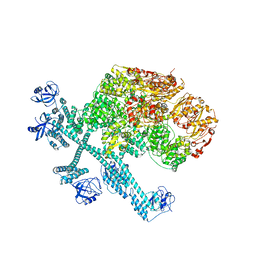 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
