8D8K
 
 | | Yeast mitochondrial small subunit assembly intermediate (State 2) | | Descriptor: | 15S ribosomal RNA, 37S ribosomal protein MRP1, mitochondrial, ... | | Authors: | Burnside, C, Harper, N.J, Klinge, S. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
7P8P
 
 | | Crystal structure of Fhit covalently bound to a nucleotide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bis(5'-adenosyl)-triphosphatase, SODIUM ION, ... | | Authors: | Herzog, D, Missun, M, Diederichs, K, Marx, A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Chemical Proteomics of the Tumor Suppressor Fhit Covalently Bound to the Cofactor Ap 3 A Elucidates Its Inhibitory Action on Translation.
J.Am.Chem.Soc., 144, 2022
|
|
5J20
 
 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J64
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(2,4-dihydroxyphenyl)-4-(2-fluorophenyl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
7V6T
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Crystal structure of bacterial DD-endopeptidase
To be published
|
|
8Z53
 
 | | Arabidopsis Dwarf14 (AtD14) co-crystallized in the presence of Zaxinone | | Descriptor: | (7E)-6-methyl-8-[(4S)-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octa-3,5,7-trien-2-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arold, S.T, Hameed, U.F.S. | | Deposit date: | 2024-04-18 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Arabidopsis Dwarf14 (AtD14) co-crystallized in the presence of Zaxinone
To Be Published
|
|
8FOW
 
 | | Ternary complex of CDK2 with small molecule ligands TW8672 and Dinaciclib | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
6NOB
 
 | |
4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
6QTJ
 
 | | Crystal structure of human CDK8/CYCC in complex with BI 919811 | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, ~{N},~{N}-dimethyl-2-[4-[4-(2,6-naphthyridin-4-yl)phenyl]pyrazol-1-yl]ethanamide | | Authors: | Boettcher, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
8SIV
 
 | | Structure of Compound 2 bound to the CHK1 10-point mutant | | Descriptor: | N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)cyclopropanecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
7PHF
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
6NTC
 
 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 1 of CRAF Kinase protein | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
7PHE
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
6OP9
 
 | | HER3 pseudokinase domain bound to bosutinib | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Littlefield, P, Agnew, C, Jura, N. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Targetable HER3 functions driving tumorigenic signaling in HER2-amplified cancers.
Cell Rep, 38, 2022
|
|
7RT1
 
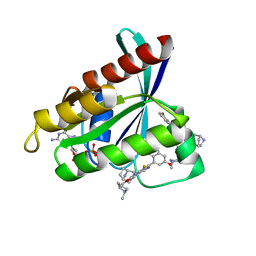 | | Crystal Structure of KRAS G12D with compound 15 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
6Y8L
 
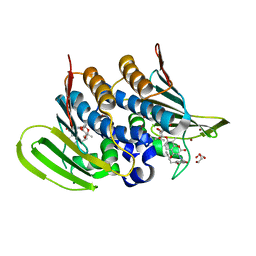 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
7RT5
 
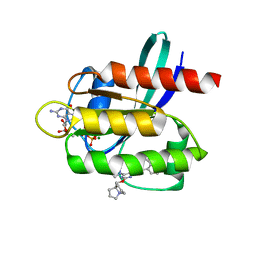 | | Crystal structure of KRAS G12D with compound 36 (4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine) bound | | Descriptor: | 4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
8A5U
 
 | |
1O3Z
 
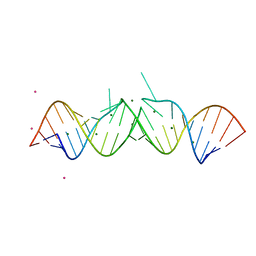 | | HIV-1 DIS(MAL) DUPLEX RU HEXAMINE-SOAKED | | Descriptor: | HIV-1 DIS(MAL) GENOMIC RNA, MAGNESIUM ION, RUTHENIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2003-05-16 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes.
Nucleic Acids Res., 31, 2003
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
8DH1
 
 | | T7 RNA polymerase elongation complex with unnatural base dDs-PaTP pair | | Descriptor: | 1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-ribofuranosyl}-1H-pyrrole-2-carbaldehyde, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2022-06-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase.
Nat Commun, 14, 2023
|
|
6YG1
 
 | | Crystal structure of MKK7 (MAP2K7) in an active state, allosterically triggered by the N-terminal helix | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, SODIUM ION | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC), Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
