4ZRX
 
 | |
6OCQ
 
 | | Crystal structure of RIP1 kinase in complex with a pyrrolidine | | Descriptor: | 1-[(2S)-2-(3-fluorophenyl)pyrrolidin-1-yl]-2,2-dimethylpropan-1-one, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
9LS8
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium at 1.22 A | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Arora, A. | | Deposit date: | 2025-02-04 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Characterization of structure of peptidyl-tRNA hydrolase from Enterococcus faecium and its inhibition by a pyrrolinone compound.
Int J Biol Macromol, 275, 2024
|
|
4ZIM
 
 | |
6NLG
 
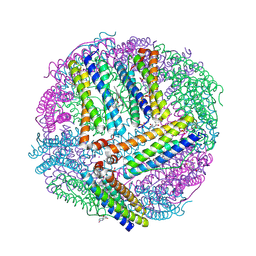 | | 1.50 A resolution structure of BfrB (C89S/K96C) from Pseudomonas aeruginosa in complex with a small molecule fragment (analog 1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-hydroxy-1H-isoindole-1,3(2H)-dione, Bacterioferritin, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6O9P
 
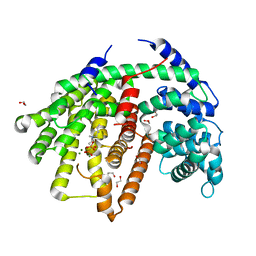 | | Wild-type SaSQS1 Complexed with Ibandronate | | Descriptor: | 1,2-ETHANEDIOL, IBANDRONATE, MAGNESIUM ION, ... | | Authors: | Blank, P.N, Christianson, D.W. | | Deposit date: | 2019-03-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained [3.1.0] Bridged-Bicyclic Product.
Acs Chem.Biol., 14, 2019
|
|
6ODM
 
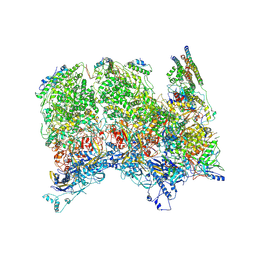 | | Herpes simplex virus type 1 (HSV-1) portal vertex-adjacent capsid/CATC, asymmetric unit | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
7AP4
 
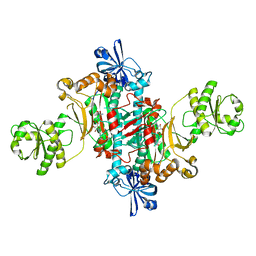 | | Thermus thermophilus Aspartyl-tRNA Synthetase in Complex with Compound AspS7HMDDA | | Descriptor: | (3~{S})-3-azanyl-4-[[(2~{R},3~{S},4~{R},5~{R})-5-[7-azanyl-5-(hydroxymethyl)benzimidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxysulfonylamino]-4-oxidanylidene-butanoic acid, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
9BUG
 
 | |
8RSN
 
 | |
9BEW
 
 | |
5XXH
 
 | | Crystal Structure Analysis of the CBP | | Descriptor: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
6O5A
 
 | |
6GQD
 
 | | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Fairhead, M, Strain-Damerell, C, Kopec, J, Bezerra, G.A, Zhang, M, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L
To Be Published
|
|
6NVU
 
 | | Crystal structure of TLA-1 extended spectrum Beta-lactamase in complex with Clavulanic Acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 2,5-bis(2-hydroxyethyl)-1,3-oxazole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Rudino-Pinera, E, Cifuentes-Castro, V.H, Rodriguez-Almazan, C. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of ESBL TLA-1 in complex with clavulanic acid reveals a second acylation site.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
8VQT
 
 | |
9BIO
 
 | |
6QYP
 
 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8A9T
 
 | | Tubulin-[1,2]oxazoloisoindole-1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
3F2L
 
 | | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Frolow, F, Freeman, A, Wine, Y, Eppel, A, Shanzer, M, Stempler, S. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus
To be Published
|
|
6VN0
 
 | | BG505 SOSIP.v4.1 in complex with rhesus macaque Fab RM20F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Cottrell, C.A, Shin, M, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
8Q5L
 
 | |
7ZUD
 
 | | Crystal structure of HIV-1 capsid IP6-CPSF6 complex | | Descriptor: | Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6, INOSITOL HEXAKISPHOSPHATE | | Authors: | Nicastro, G, Taylor, I.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
J.Mol.Biol., 434, 2022
|
|
8YLE
 
 | | Crystal structure of Werner syndrome helicase complexed with AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Yang, Y, Fu, L, Sun, X, Cheng, H, Chen, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of werner syndrome helicase complexed with AMP-PCP at 1.86 Angstroms resolution.
To Be Published
|
|
