8CTJ
 
 | | Cryo-EM structure of TMEM87A | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Transmembrane protein 87A | | Authors: | Hoel, C.M, Zhang, L, Brohawn, S.G. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.74 Å) | | Cite: | Structure of the GOLD-domain seven-transmembrane helix protein family member TMEM87A.
Elife, 11, 2022
|
|
8WS1
 
 | | Crystal structure of human NEK7 D161N mutant | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase Nek7 | | Authors: | Bijpuria, S, Kanavalli, M, Subbiah, R, Mollard, A, Bearss, D. | | Deposit date: | 2023-10-16 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human NEK7 D161N mutant
To Be Published
|
|
8W25
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament bound to compound, TDI-2804 (focused refinement map). | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Poly [ADP-ribose] polymerase tankyrase-2, N-{2-[4-(2-hydroxypropan-2-yl)phenyl]-4-oxo-1,4-dihydroquinazolin-7-yl}-4-methoxy-6-phenylpyridine-3-carboxamide, ZINC ION | | Authors: | Malone, B.F, Zimmerman, J.L, Dow, L.E, Hite, R.K. | | Deposit date: | 2024-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | A potent and selective TNKS2 inhibitor for tumor-selective WNT suppression.
Biorxiv, 2025
|
|
8DDL
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Apo Structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ORF1a polyprotein, ... | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
9AVJ
 
 | | PS3 F1 ATPase Wild type | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2024-03-03 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The molecular structure of an axle-less F 1 -ATPase.
Biochim Biophys Acta Bioenerg, 1866, 2024
|
|
8E1A
 
 | | Structure-based study to overcome cross-reactivity of novel androgen receptor inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-fluoro-2-methoxyphenyl)-1,3-thiazol-2-yl]morpholine, Androgen receptor | | Authors: | Lallous, N, Li, H, Radaeva, M, Dalal, K, Leblanc, E, Ban, F, Ciesielski, F, Chow, B, Morin, M, Singh, K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Cells, 11, 2022
|
|
8VGS
 
 | |
5NEP
 
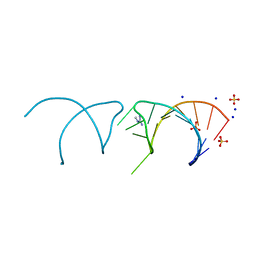 | | The structure of the G. violaceus guanidine II riboswitch P1 stem-loop with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, RNA (5'-R(*GP*GP*UP*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*AP*(CBV)P*C)-3'), SODIUM ION, ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
8X72
 
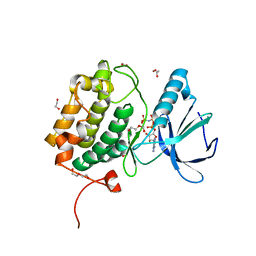 | | The Crystal Structure of PLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wu, B. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of PLK1 from Biortus.
To Be Published
|
|
7S7W
 
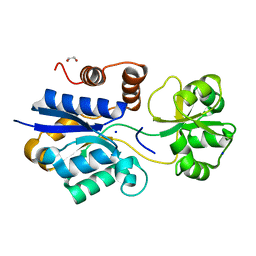 | | Crystal structure of iNicSnFR3a Nicotine Sensor precursor binding protein | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, iNicSnFR 3.0 Fluorescent Nicotine Sensor precursor binding protein | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
1NC2
 
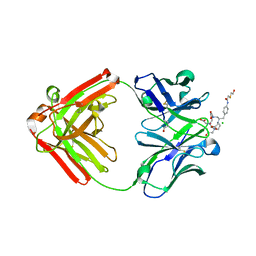 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Y-DOTA | | Descriptor: | (S)-2-(4-(2-(2-HYDROXYETHYLTHIO)-ACETAMIDO)-BENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
6NKG
 
 | | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Lip_vut5, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome.
To Be Published
|
|
7B0M
 
 | | Sugar transaminase from a metagenome collected from troll oil field production water | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sugar aminotransferase, ... | | Authors: | Littlechild, J.A, De Rose, S.A, Isupov, M.N, Sayer, C, Karki, S. | | Deposit date: | 2020-11-20 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Sugar transaminases from hot environments
To Be Published
|
|
6NCG
 
 | | Crystal Structure of Human Vaccinia-related kinase 2 (VRK-2) bound to pyridin-benzenesulfonamide inhibitor | | Descriptor: | 4-[6-amino-5-(3,5-difluoro-4-hydroxyphenyl)pyridin-3-yl]benzene-1-sulfonamide, SULFATE ION, Serine/threonine-protein kinase VRK2 | | Authors: | dos Reis, C.V, Chiodi, C.G, de Souza, G.P, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Counago, R.M, Massirer, K.B, Elkins, J.M, Arruda, P, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Vaccinia-related kinase 2 (VRK-2) bound to pyridin-benzenesulfonamide inhibitor
To Be Published
|
|
8XOX
 
 | | The Crystal Structure of FAK2 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide, Protein-tyrosine kinase 2-beta | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of FAK2 from Biortus.
To Be Published
|
|
8E2R
 
 | | Crystal structure of TadAC-1.14 | | Descriptor: | GLYCEROL, ZINC ION, tRNA-specific adenosine deaminase 1.14 | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
6WO0
 
 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | Descriptor: | GLYCEROL, Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
8XEJ
 
 | | Cryo-EM structure of human XKR8-basigin complex in lipid nanodisc | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab heavy chain, Fab light chain, ... | | Authors: | Sakuragi, T.S, Kanai, R.K, Kikkawa, M.K, Toyoshima, C.T, Nagata, S.N. | | Deposit date: | 2023-12-12 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The role of the C-terminal tail region as a plug to regulate XKR8 lipid scramblase.
J.Biol.Chem., 300, 2024
|
|
5X6Y
 
 | |
9AZW
 
 | | Crystal structure of PDC-88 beta-lactamase in complex with taniborbactam | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2024-03-11 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of taniborbactam inhibition of the cefepime-hydrolyzing, partial R2-loop deletion Pseudomonas -derived cephalosporinase variant PDC-88.
Antimicrob.Agents Chemother., 69, 2025
|
|
6XMI
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Fab Heavy chain, Fab Light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7ALZ
 
 | | GqqA- a novel type of quorum quenching acylases | | Descriptor: | PHENYLALANINE, Prephenate dehydratase | | Authors: | Werner, N, Betzel, C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Komagataeibacter europaeus GqqA is the prototype of a novel bifunctional N-Acyl-homoserine lactone acylase with prephenate dehydratase activity.
Sci Rep, 11, 2021
|
|
6XD5
 
 | | Apo KPC-2 N170A mutant at 1.20 A | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, SULFATE ION | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
9B1B
 
 | | EV-D68 in complex with inhibitor Jun11-78-7 | | Descriptor: | 4-[(propan-2-yl)oxy]-N-{(4M)-4-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-4-yl]quinolin-8-yl}benzamide, Capsid protein VP1, Capsid protein VP4, ... | | Authors: | Klose, T, Kuhn, R.J, Jun, W, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-03-13 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Design of enterovirus D68 capsid inhibitors with in vivo antiviral efficacy
To Be Published
|
|
9B18
 
 | |
