8DPQ
 
 | | Beta-lactamase CTX-M-14 N170A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Lu, S, Neetu, N, Palzkill, T. | | Deposit date: | 2022-07-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
6NFK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
8OTQ
 
 | | Cryo-EM structure of Strongylocentrotus purpuratus sperm-specific Na+/H+ exchanger SLC9C1 in GDN | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PALMITIC ACID, Sperm-specific sodium proton exchanger | | Authors: | Yeo, H, Mehta, V, Gulati, A, Drew, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure and electromechanical coupling of a voltage-gated Na + /H + exchanger.
Nature, 623, 2023
|
|
5LXZ
 
 | |
8VA0
 
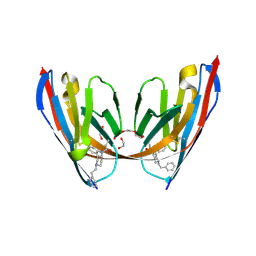 | | X-ray crystal structure of JGFN4 N76D complexed with fentanyl in dimer form | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Shi, K, Moller, N, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
4YCC
 
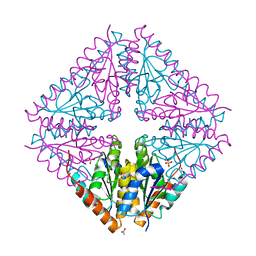 | |
8WT0
 
 | | The toxin-antitoxin complex Fic-1-AntF is a deAMPylase that regulates the activity of DNA gyrase | | Descriptor: | Cell filamentation protein Fic, D(-)-TARTARIC ACID, DUF2559 domain-containing protein, ... | | Authors: | Chen, F.R, Guo, L.W, Lu, C.H, Jiang, W.J, Luo, Z.Q, Liu, J.F, Zhang, L.Q. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The toxin-antitoxin complex Fic-1-AntF is a deAMPylase that regulates the activity of DNA gyrase
To Be Published
|
|
8P1X
 
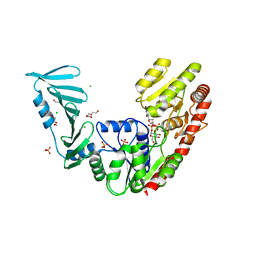 | | TarM(Se)_G117R-UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2023-05-13 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
9FHE
 
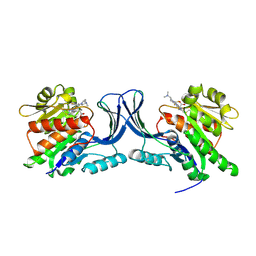 | | hKHK-C in complex with BI-9787 (pH 5.5) | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
9FCS
 
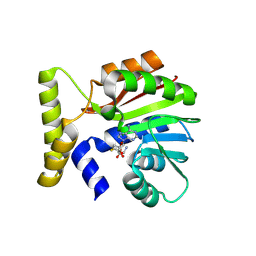 | | CysG(N-16)-H122N mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
6E1U
 
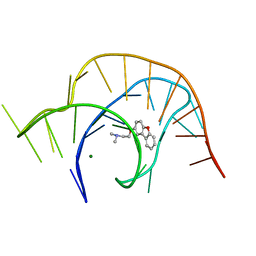 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 2: 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine, MAGNESIUM ION, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
9FLT
 
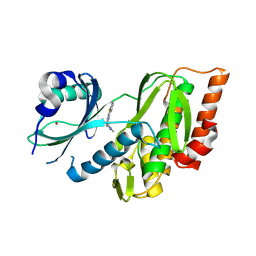 | | Crystal structure of human Haspin (GSG2) kinase bound to chemical probe MU1920 | | Descriptor: | NICKEL (II) ION, Serine/threonine-protein kinase haspin, ~{N}-(1,4-dimethylpyrazol-3-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridin-5-amine | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8EO5
 
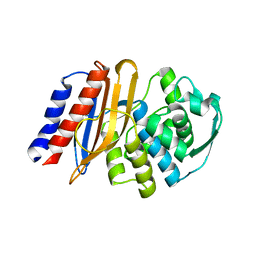 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
9H8I
 
 | | Crystallization of B. licheniformis levanase | | Descriptor: | 1,2-ETHANEDIOL, BICINE, CALCIUM ION, ... | | Authors: | Carr, S, Cruz-Migoni, A, Porras-Dominguez, J.R, Van den Ende, W, Lopez-Munguia Canales, A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Understanding the Endo- and Exo-mechanisms Involved in the Enzymatic Hydrolysis of Levan and Inulin Polymers.
J.Agric.Food Chem., 73, 2025
|
|
9HY3
 
 | | sc-4E (scFv derived from the mAb 4E1 against CD93) crystallized at pH 5.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural and antigen-binding surface definition of an anti-CD93 monoclonal antibody for the treatment of degenerative vascular eye diseases.
Int.J.Biol.Macromol., 309, 2025
|
|
9HY1
 
 | | sc-4E (scFv derived from the mAb 4E1 against CD93) crystallized at pH 7.5 (dimeric state from gel filtration) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and antigen-binding surface definition of an anti-CD93 monoclonal antibody for the treatment of degenerative vascular eye diseases.
Int.J.Biol.Macromol., 309, 2025
|
|
6ZXY
 
 | |
8DMQ
 
 | |
7A4L
 
 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | Descriptor: | IRON/SULFUR CLUSTER, PioC | | Authors: | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
7AE6
 
 | | Crystal structure of di-AMPylated HEPN(R102A) toxin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7NZD
 
 | | Fourth SH3 domain of POSH (Plenty of SH3 Domains protein) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase SH3RF1 | | Authors: | Palencia, A, Bessa, L.M, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
1GLI
 
 | |
7OFY
 
 | | Crystal structure of SQ binding protein from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | 1,2-ETHANEDIOL, Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Sharma, M, Goddard-Borger, E.D, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4Y9D
 
 | | Crystal structure of LigD in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
8C54
 
 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
