5Y3J
 
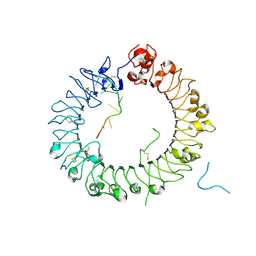 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and TCGCAC DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2017-07-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
5Y3K
 
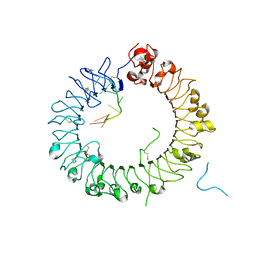 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and GCGCAC DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2017-07-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
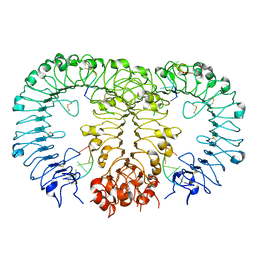
5Y3M
 
 | |
6Q0C
 
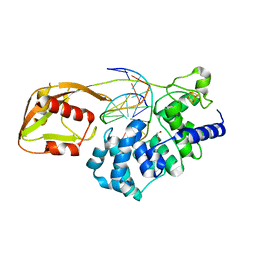 | | MutY adenine glycosylase bound to DNA containing a transition state analog (1N) paired with undamaged dG | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, CALCIUM ION, ... | | Authors: | O'Shea Murray, V.L, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2019-08-01 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY.
Acs Chem.Biol., 15, 2020
|
|
5Y3L
 
 | | Crystal structure of horse TLR9 in complex with two DNAs (CpG DNA and CCGCAC DNA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*AP*GP*GP*CP*GP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2017-07-29 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Immunity, 48, 2018
|
|
3BTY
 
 | | Crystal structure of human ABH2 bound to dsDNA containing 1meA through cross-linking away from active site | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DGP*DTP*DTP*DAP*DTP*DAP*DCP*DA)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
7LXH
 
 | | Bacillus cereus DNA glycosylase AlkD bound to a CC1065-adenine nucleobase adduct and DNA containing an abasic site | | Descriptor: | 7-{7-[(1R)-1-{[(4P)-6-amino-3H-purin-3-yl]methyl}-5-hydroxy-8-methyl-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carbonyl]-4-hydroxy-5-methoxy-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carbonyl}-4-hydroxy-5-methoxy-1,6-dihydropyrrolo[3,2-e]indole-3(2H)-carboxamide, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*AP*(ORP)P*GP*GP*C)-3'), ... | | Authors: | Mullins, E.A, Eichman, B.F. | | Deposit date: | 2021-03-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Structural evolution of a DNA repair self-resistance mechanism targeting genotoxic secondary metabolites.
Nat Commun, 12, 2021
|
|
7LXJ
 
 | |
6G40
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH9525 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2014-04-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
1VTY
 
 | | Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs | | Descriptor: | AMINO GROUP, DNA (5'-D(*CP*(NH2)AP*CP*GP*TP*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Wang, A.H.-J, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs.
J. Biomol. Struct. Dyn., 4, 1986
|
|
8WU5
 
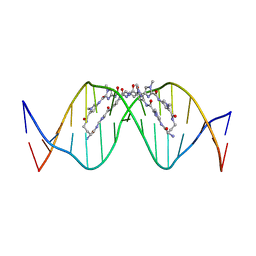 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | Descriptor: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | Authors: | Abe, K, Takeda, K, Sugiyama, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
1EWR
 
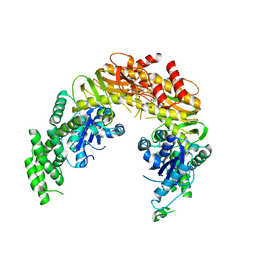 | | CRYSTAL STRUCTURE OF TAQ MUTS | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Obmolova, G, Ban, C, Hsieh, P, Yang, W. | | Deposit date: | 2000-04-26 | | Release date: | 2000-10-23 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA.
Nature, 407, 2000
|
|
3MGI
 
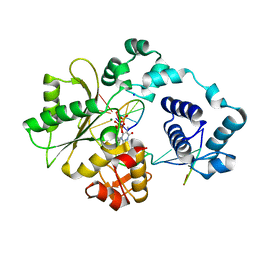 | | Ternary complex of a DNA polymerase lambda loop mutant | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA, DNA (5'-D(*CP*AP*GP*TP*AP*T)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Zhou, R.Z, Povirk, L.F, Kunkel, T. | | Deposit date: | 2010-04-06 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Loop 1 modulates the fidelity of DNA polymerase lambda
Nucleic Acids Res., 38, 2010
|
|
6U16
 
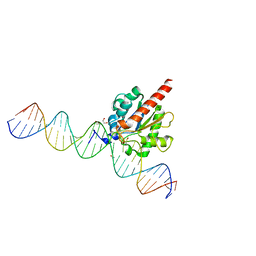 | | Human thymine DNA glycosylase N140A mutant bound to DNA with 5-carboxyl-dC substrate | | Descriptor: | 1,2-ETHANEDIOL, DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2019-08-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Excision of 5-Carboxylcytosine by Thymine DNA Glycosylase.
J.Am.Chem.Soc., 141, 2019
|
|
1HUO
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
6GAV
 
 | | Extremely 'open' clamp structure of DNA gyrase: role of the Corynebacteriales GyrB specific insert | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B,DNA gyrase subunit A | | Authors: | Petrella, S, Capton, E, Alzari, P.M, Aubry, A, MAyer, C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a Corynebacteriales GyrB-Specific Insert in ATPase Activity.
Structure, 27, 2019
|
|
3V72
 
 | | Crystal Structure of Rat DNA polymerase beta Mutator E295K: Enzyme-dsDNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(P*AP*AP*AP*CP*TP*CP*AP*CP*AP*T)-3', DNA 5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3', ... | | Authors: | Gridley, C.L, Jaeger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Unfavorable Electrostatic and Steric Interactions in DNA Polymerase beta E295K Mutant Interfere with the Enzyme s Pathway
J.Am.Chem.Soc., 134, 2012
|
|
3BIE
 
 | |
3BTZ
 
 | | Crystal structure of human ABH2 cross-linked to dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*AP*GP*GP*TP*GP*AP*(2YR)P*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DTP*DTP*DAP*DTP*DCP*DAP*DCP*DC)-3') | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BUC
 
 | | X-ray structure of human ABH2 bound to dsDNA with Mn(II) and 2KG | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*DCP*DTP*DGP*DTP*DAP*DTP*(MA7)P*DAP*DCP*DTP*DGP*DCP*DG)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3ZOR
 
 | | Structure of BsUDG | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Banos-Sanz, J.I, Mojardin, L, Sanz-Aparicio, J, Gonzalez, B, Salas, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure and Functional Insights Into Uracil-DNA Glycosylase Inhibition by Phage Phi29 DNA Mimic Protein P56
Nucleic Acids Res., 41, 2013
|
|
7V6W
 
 | | Crystal structure of heterohexameric Sa2YoeB-Sa2YefM complex bound to 26bp-DNA | | Descriptor: | Antitoxin, DNA (25-MER), DNA (26-MER), ... | | Authors: | Xue, L, Khan, M.H, Yue, J. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The two paralogous copies of the YoeB-YefM toxin-antitoxin module in Staphylococcus aureus differ in DNA binding and recognition patterns.
J.Biol.Chem., 298, 2022
|
|
5K83
 
 | | Crystal Structure of a Primate APOBEC3G N-Domain, in Complex with ssDNA | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
