4UOF
 
 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with dADP | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from the Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4UOG
 
 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with dCDP | | Descriptor: | DEOXYCYTIDINE DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
2O3V
 
 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33 | | Descriptor: | (2S,3R,4R,5S,6R)-3-AMINO-4-({[(2S,3R,4R,5S,6R)-3-AMINO-2-{[(1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYL]OXY}-5-HYDROXY-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-4-YL]OXY}METHOXY)-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-2,5-DIOL, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
6I75
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,5,6-tetrakis(fluoranyl)-4-oxidanyl-phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
4JSU
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:3a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome
J.Med.Chem., 56, 2013
|
|
6I74
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 1 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6OI2
 
 | | Crystal structure of human WDR5 in complex with symmetric dimethyl-L-arginine | | Descriptor: | GLYCEROL, N3, N4-DIMETHYLARGININE, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
4JSQ
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:4e | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome
J.Med.Chem., 56, 2013
|
|
5J1T
 
 | | TorsinAdeltaE-LULL1 complex, H. sapiens, bound to VHH-BS2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Demircioglu, F.E, Schwartz, T.U. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia.
Elife, 5, 2016
|
|
5J31
 
 | | Crystal structure of 14-3-3zeta in complex with an alkyne cross-linked cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Exoenzyme S | | Authors: | Wallraven, K, Cromm, P, Bier, D, Glas, A, Grossmann, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constraining an Irregular Peptide Secondary Structure through Ring-Closing Alkyne Metathesis.
Chembiochem, 17, 2016
|
|
6OWH
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 92 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-05-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
2O3W
 
 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site in presence of paromomycin | | Descriptor: | PAROMOMYCIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
7VRL
 
 | | Solution structure of Rbfox RRM bound to a non-cognate RNA | | Descriptor: | RNA (5'-R(*UP*GP*CP*AP*UP*AP*U)-3'), RNA binding protein fox-1 homolog 1 | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2021-10-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two distinct binding modes provide the RNA-binding protein RbFox with extraordinary sequence specificity.
Nat Commun, 14, 2023
|
|
5OPA
 
 | | The crystal structure of P450 CYP121 in complex with lead compound 7b | | Descriptor: | 3-(4-fluorophenyl)-4-(imidazol-1-ylmethyl)-1-phenyl-pyrazole, DI(HYDROXYETHYL)ETHER, Mycocyclosin synthase, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
4UDD
 
 | | GR in complex with desisobutyrylciclesonide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESISOBUYTYRYL CICLESONIDE, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
6P5U
 
 | | Structure of an enoyl-CoA hydratase/aldolase isolated from a lignin-degrading consortium | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Enoyl-CoA hydratase | | Authors: | Liberato, M.V, Squina, F.M. | | Deposit date: | 2019-05-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a prokaryotic feruloyl-CoA hydratase-lyase from a lignin-degrading consortium with high oligomerization stability under extreme pHs.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
4UDC
 
 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDB
 
 | | MR in complex with desisobutyrylciclesonide | | Descriptor: | DESISOBUYTYRYL CICLESONIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, JellesmarkJensen, T, Cavallin, A, Nilsson, E, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDA
 
 | | MR in complex with dexamethasone | | Descriptor: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UOH
 
 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Guevara-Hernandez, E, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
6HIV
 
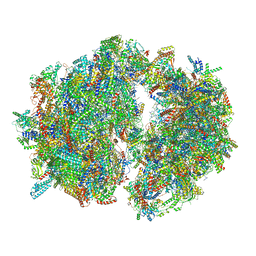 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the complete mitoribosome | | Descriptor: | 12S rRNA, 50S ribosomal protein L13, putative, ... | | Authors: | Ramrath, D.J.F, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boerhringer, D, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-07 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
4GR6
 
 | | Crystal structure of AtRbcX2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, AtRbcX2 | | Authors: | Grudnik, P, Golik, P, Kolesinski, P, Dubin, G, Szczepaniak, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into eukaryotic Rubisco assembly - Crystal structures of RbcX chaperones from Arabidopsis thaliana.
Biochim.Biophys.Acta, 1830, 2013
|
|
7E7L
 
 | | The crystal structure of arylacetate decarboxylase from Olsenella scatoligenes. | | Descriptor: | 4-HYDROXYPHENYLACETATE, Hydroxyphenylacetic acid decarboxylase | | Authors: | Lu, Q, Duan, Y, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The Glycyl Radical Enzyme Arylacetate Decarboxylase from Olsenella scatoligenes
Acs Catalysis, 11, 2021
|
|
5LOX
 
 | |
2O3Y
 
 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site in Presence of Paromamine Derivative NB30 | | Descriptor: | RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3'), SPERMINE | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
