7R0A
 
 | | Structure of sarin phosphonylated acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R02
 
 | | Mus musculus acetylcholinesterase in complex with N-(3-(diethylamino)propyl)-4-methyl-3-nitrobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
6T25
 
 | | Cryo-EM structure of phalloidin-Alexa Flour-546-stabilized F-actin (copolymerized) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
7R1O
 
 | | p62-ZZ domain of the human sequestosome in complex with dusquetide | | Descriptor: | Dusquetide, Sequestosome-1, ZINC ION | | Authors: | Hakansson, M, Hansson, M, Logan, D.T, Rozek, A, Donini, O. | | Deposit date: | 2022-02-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Dusquetide modulates innate immune response through binding to p62.
Structure, 30, 2022
|
|
6T23
 
 | | Cryo-EM structure of jasplakinolide-stabilized F-actin (aged) | | Descriptor: | (4~{R},7~{R},10~{S},13~{S},15~{E},19~{S})-10-(4-azanylbutyl)-4-(4-hydroxyphenyl)-7-(1~{H}-indol-3-ylmethyl)-8,13,15,19-tetramethyl-1-oxa-5,8,11-triazacyclononadec-15-ene-2,6,9,12-tetrone, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
6SYT
 
 | | Structure of the SMG1-SMG8-SMG9 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gat, Y, Schuller, J.M, Conti, E. | | Deposit date: | 2019-10-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | InsP6binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6SHD
 
 | |
3BX5
 
 | | P38 alpha map kinase complexed with BMS-640994 | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-methyl-5-(methylcarbamoyl)phenyl]-2-{[(1R)-1-methylpropyl]amino}-1,3-thiazole-5-carboxamide | | Authors: | Sack, J.S. | | Deposit date: | 2008-01-11 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The discovery of (R)-2-(sec-butylamino)-N-(2-methyl-5-(methylcarbamoyl)phenyl) thiazole-5-carboxamide (BMS-640994)-A potent and efficacious p38alpha MAP kinase inhibitor
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4MIH
 
 | | Pyranose 2-oxidase from Phanerochaete chrysosporium, recombinant H158A mutant | | Descriptor: | 3-deoxy-3-fluoro-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Hassan, N, Tan, T.C, Spadiut, O, Pisanelli, I, Fusco, L, Haltrich, D, Peterbauer, C, Divne, C. | | Deposit date: | 2013-08-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Phanerochaete chrysosporium pyranose 2-oxidase suggest that the N-terminus acts as a propeptide that assists in homotetramer assembly.
FEBS Open Bio, 3, 2013
|
|
3BPB
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase H162G adduct with S-methyl-L-thiocitrulline | | Descriptor: | N~5~-[(E)-imino(methylsulfanyl)methyl]-L-ornithine, dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Linsky, T.W, Stone, E.M, Fast, W, Robertus, J.D. | | Deposit date: | 2007-12-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Promiscuous partitioning of a covalent intermediate common in the pentein superfamily.
Chem.Biol., 15, 2008
|
|
3BV2
 
 | | Morpholino pyrrolotriazine P38 Alpha map kinase inhibitor compound 30 | | Descriptor: | 5-methyl-4-[(2-methyl-5-{[(2-morpholin-4-ylpyridin-4-yl)carbonyl]amino}phenyl)amino]-N-(1-phenylethenyl)pyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and SAR of new pyrrolo[2,1-f][1,2,4]triazines as potent p38 alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4HWJ
 
 | |
2GAS
 
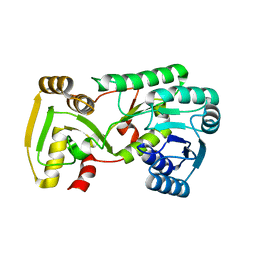 | | Crystal Structure of Isoflavone Reductase | | Descriptor: | isoflavone reductase | | Authors: | Wang, X, He, X, Lin, J, Shao, H, Chang, Z, Dixon, R.A. | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Isoflavone Reductase from Alfalfa (Medicago sativa L.)
J.Mol.Biol., 358, 2006
|
|
4IY5
 
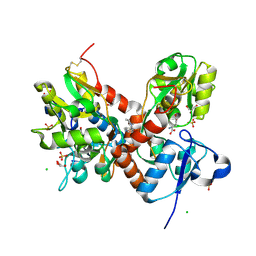 | | Crystal structure of the glua2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and CX516 at 2.0 A resolution | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-01-28 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4NWD
 
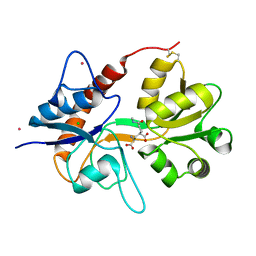 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4IIA
 
 | |
4M4L
 
 | | The structure of Cu T6 bovine insulin | | Descriptor: | COPPER (II) ION, Insulin | | Authors: | Frankaer, C.G, Harris, P, Stahl, K. | | Deposit date: | 2013-08-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Towards accurate structural characterization of metal centres in protein crystals: the structures of Ni and Cu T6 bovine insulin derivatives.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4H9G
 
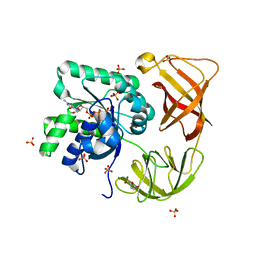 | | Probing EF-Tu with a very small brominated fragment library identifies the CCA pocket | | Descriptor: | 5-bromofuran-2-carboxylic acid, AMMONIUM ION, Elongation factor Tu-A, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R.H, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2012-09-24 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4O6K
 
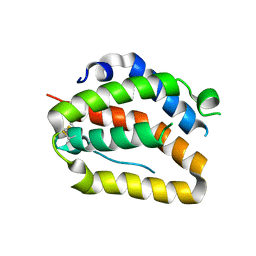 | | The crystal structure of zebrafish IL-22 | | Descriptor: | Interleukin 22 | | Authors: | Siupka, P, Hamming, O.J, Fretaud, M, Luftalla, G, Levraud, J.P, Hartmann, R. | | Deposit date: | 2013-12-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of zebrafish IL-22 reveals an evolutionary, conserved structure highly similar to that of human IL-22.
Genes Immun., 15, 2014
|
|
4N07
 
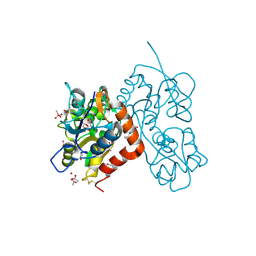 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold.
J.Med.Chem., 56, 2013
|
|
4JHI
 
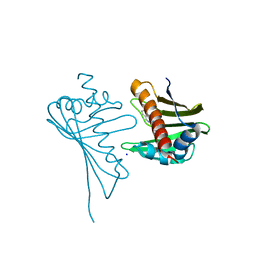 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-benzyladenine | | Descriptor: | MtN13 protein, N-BENZYL-9H-PURIN-6-AMINE, SODIUM ION | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4M4M
 
 | | The structure of Ni T6 bovine insulin | | Descriptor: | Insulin, NICKEL (II) ION | | Authors: | Frankaer, C.G, Harris, P, Stahl, K. | | Deposit date: | 2013-08-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards accurate structural characterization of metal centres in protein crystals: the structures of Ni and Cu T6 bovine insulin derivatives.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LP4
 
 | |
4IY6
 
 | | Crystal structure of the GLUA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and ME-CX516 at 1.72 A resolution | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-01-28 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J0N
 
 | | Crystal structure of a manganese dependent isatin hydrolase | | Descriptor: | CALCIUM ION, Isatin hydrolase B, MANGANESE (II) ION, ... | | Authors: | Bjerregaard-Andersen, K, Sommer, T, Jensen, J.K, Jochimsen, B, Etzerodt, M, Morth, J.P. | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A proton wire and water channel revealed in the crystal structure of isatin hydrolase.
J.Biol.Chem., 289, 2014
|
|
