8VJP
 
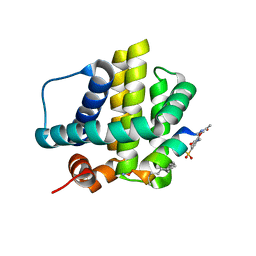 | | Histidine-covalent stapled alpha-helical peptide (155H1) targeting hMcl-1 | | Descriptor: | (4Z)-oct-4-en-1-ol, (S~1~R)-3-carbamoyl-4-methoxybenzene-1-sulfinic acid, Histidine-covalent stapled alpha-helical peptide, ... | | Authors: | Muzzarelli, K.M, Assar, Z, Alboreggia, G, Pellecchia, M. | | Deposit date: | 2024-01-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Histidine-Covalent Stapled Alpha-Helical Peptides Targeting hMcl-1.
J.Med.Chem., 67, 2024
|
|
7R3O
 
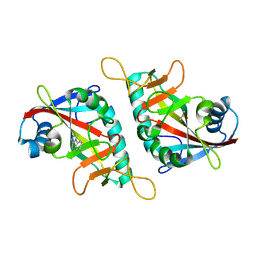 | | PARP15 catalytic domain in complex with OUL40 | | Descriptor: | 6-methyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7R4A
 
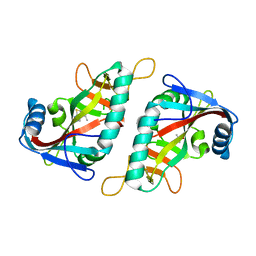 | | PARP15 catalytic domain in complex with OUL188 | | Descriptor: | 6,8-dimethyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Murthy, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7RSA
 
 | |
6NOL
 
 | |
6Y8I
 
 | |
6XKR
 
 | | Structure of Sasanlimab Fab in complex with PD-1 | | Descriptor: | GLYCEROL, Programmed cell death protein 1, Sasanlimab Fab Heavy chain, ... | | Authors: | Kimberlin, C.R, Chin, S.M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Pharmacologic Properties and Preclinical Activity of Sasanlimab, A High-affinity Engineered Anti-Human PD-1 Antibody.
Mol.Cancer Ther., 19, 2020
|
|
6PBM
 
 | | Pseudopaline Dehydrogenase with NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline Dehydrogenase | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6Z5F
 
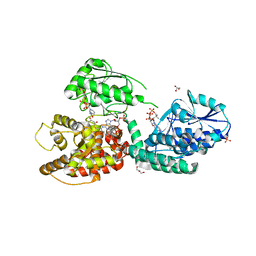 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH 3-KETODECANOYL-COA AND OXIDISED NICOTINAMIDE ADENINE DINUCLEOTIDE | | Descriptor: | 3-KETO-DECANOYL-COA, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic binding studies of rat peroxisomal multifunctional enzyme type 1 with 3-ketodecanoyl-CoA: capturing active and inactive states of its hydratase and dehydrogenase catalytic sites.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5JRS
 
 | |
9I4F
 
 | | Blood Type B-converting alpha-1,3-galactosidase PpaGal from Pedobacter panaciterrae in its apo form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3-galactosidase PpaGal | | Authors: | Schmoeker, O, Moeller, C, Terholsen, H, Girbardt, B, Palm, G.J, Hoppen, J, Lammers, M, Bornscheuer, U.T. | | Deposit date: | 2025-01-24 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification and Protein Engineering of Galactosidases for the Conversion of Blood Type B to Blood Type O.
Chembiochem, 26, 2025
|
|
9MLM
 
 | | Crystal structure of dihydrofolate reductase (DHFR) from the filarial nematode W. bancrofti in complex with NADPH and [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-methoxyphenyl]acetic acid (TSD10 or OED) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-methoxyphenyl]acetic acid | | Authors: | Frey, K.M, Goodey, N.M, Kwarteng, S, Wilhelm, J. | | Deposit date: | 2024-12-19 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A virtual screening strategy to repurpose antifolate compounds as W.bancrofti DHFR inhibitors.
Bioorg.Med.Chem.Lett., 129, 2025
|
|
7C7O
 
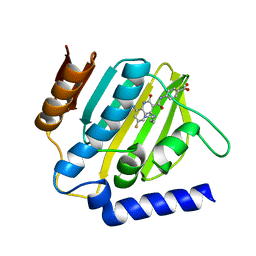 | | Crystal structure of E.coli DNA gyrase B in complex with 6-fluoro-8-(methylamino)-2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[4-(3-azanylpropylamino)-6-fluoranyl-8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Kamitani, M, Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lead optimization of 8-(methylamino)-2-oxo-1,2-dihydroquinolines as bacterial type II topoisomerase inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
5YKJ
 
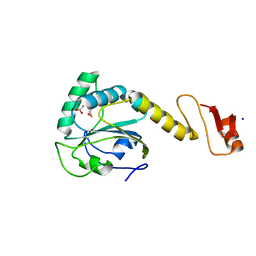 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | GLYCEROL, Peroxiredoxin PRX1, mitochondrial, ... | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-14 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
To be published
|
|
7C7N
 
 | |
6ZIB
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH ACETOACETYL-COA AND NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETOACETYL-COENZYME A, Peroxisomal bifunctional enzyme, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-06-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural enzymology studies with the substrate 3S-hydroxybutanoyl-CoA: bifunctional MFE1 is a less efficient dehydrogenase than monofunctional HAD.
Febs Open Bio, 14, 2024
|
|
6ZIC
 
 | | CRYSTAL STRUCTURE OF RAT PEROXISOMAL MULTIFUNCTIONAL ENZYME TYPE-1 (RPMFE1) COMPLEXED WITH 3S-HYDROXYBUTANOYL-COA AND NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-HYDROXYBUTANOYL-COENZYME A, GLYCEROL, ... | | Authors: | Wierenga, R.K, Sridhar, S, Kiema, T.R. | | Deposit date: | 2020-06-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural enzymology studies with the substrate 3S-hydroxybutanoyl-CoA: bifunctional MFE1 is a less efficient dehydrogenase than monofunctional HAD.
Febs Open Bio, 14, 2024
|
|
5KM6
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant Ara-A nucleoside phosphoramidate substrate complex | | Descriptor: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[2-(1~{H}-indol-3-yl)ethyl]phosphonamidic acid | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
6M95
 
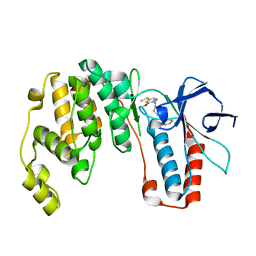 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping: compound 1 | | Descriptor: | (4-benzylpiperidin-1-yl)[2-methoxy-4-(methylsulfanyl)phenyl]methanone, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
7TEV
 
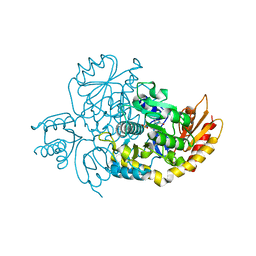 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (3S,4R)-3-amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylate | | Descriptor: | (1S,3R,4S)-3-formyl-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
9XIA
 
 | |
6XKH
 
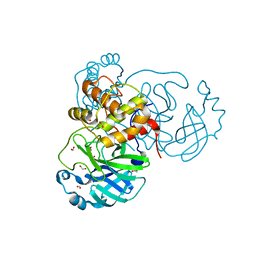 | | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine) | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, ACETATE ION, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevsky, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | THE 1.28A CRYSTAL STRUCTURE OF 3CL MAINPRO OF SARS-COV-2 WITH OXIDIZED C145 (sulfinic acid cysteine)
To Be Published
|
|
5IDT
 
 | |
9VEM
 
 | |
6XFI
 
 | | Crystal Structures of beta-1,4-N-Acetylglucosaminyltransferase 2 (POMGNT2): Structural Basis for Inherited Muscular Dystrophies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2, ... | | Authors: | Halmo, S.M, Yeh, J, Wells, L, Moremen, K.W, Lanzilotta, W.N. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of beta-1,4-N-acetylglucosaminyltransferase 2: structural basis for inherited muscular dystrophies.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
