5GTG
 
 | | Crystal structure of onion lachrymatory factor synthase (LFS) containing 1,2-propanediol | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lachrymatory-factor synthase, S-1,2-PROPANEDIOL, ... | | Authors: | Arakawa, T, Sato, Y, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2016-08-20 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase
Acs Catalysis, 10, 2020
|
|
3WN2
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylohexaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
2L1T
 
 | |
6US4
 
 | | MTH1 in complex with compound 32 | | Descriptor: | 5-(2,3-dichlorophenyl)[1,2,4]triazolo[1,5-a]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Newby, Z.E.R, Lansdon, E.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95032907 Å) | | Cite: | Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Acs Med.Chem.Lett., 11, 2020
|
|
6USA
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and GSK1-bound form | | Descriptor: | (3R,4R)-4-[4-(2-Chlorophenyl)piperazin-1-yl]-1,1-dioxothiolan-3-ol, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Allosteric inhibitors of Mycobacterium tuberculosis tryptophan synthase.
Protein Sci., 29, 2020
|
|
8XKS
 
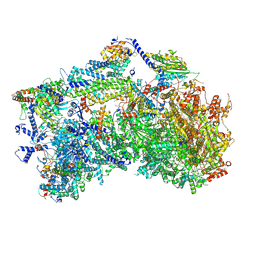 | | The cryo-EM structure of Orf2971-FtsHi motor complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 4Fe-4S ferredoxin-type domain-containing protein, ... | | Authors: | Wang, N, Li, M. | | Deposit date: | 2023-12-24 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Architecture of the ATP-driven motor for protein import into chloroplasts.
Mol Plant, 17, 2024
|
|
5GMQ
 
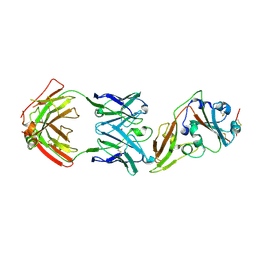 | | Structure of MERS-CoV RBD in complex with a fully human antibody MCA1 | | Descriptor: | 1,2-ETHANEDIOL, MCA1 heavy chain, MCA1 light chain, ... | | Authors: | Chen, C, Wang, J.M, Zou, T.T, Gao, X.P, Cui, S, Jin, Q. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
J. Infect. Dis., 215, 2017
|
|
3AZ8
 
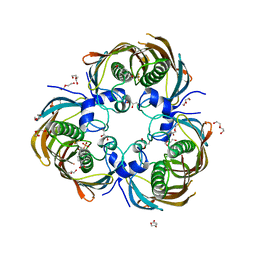 | | Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum in complex with NAS21 | | Descriptor: | 4,4,4-trifluoro-1-(4-nitrophenyl)butane-1,3-dione, Beta-hydroxyacyl-ACP dehydratase, CHLORIDE ION, ... | | Authors: | Maity, K, Venkata, B.S, Kapoor, N, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2011-05-20 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the functional and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum
J.Struct.Biol., 176, 2011
|
|
6US3
 
 | | MTH1 in complex with compound 4 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N-[5-(2,3-dimethylphenyl)-1,6-naphthyridin-7-yl]acetamide | | Authors: | Newby, Z.E.R, Lansdon, E.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47028923 Å) | | Cite: | Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Acs Med.Chem.Lett., 11, 2020
|
|
6UVJ
 
 | |
6UVP
 
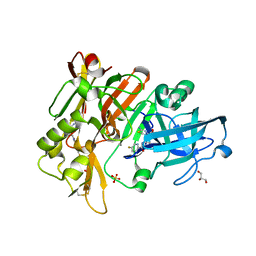 | | BACE-1 in complex with compound #3 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]cyclopropyl}-5-fluoropyridine-2-carboxamide, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
4H6B
 
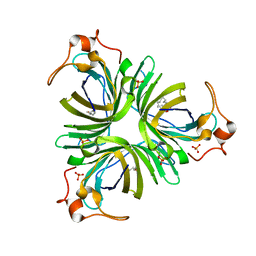 | | Structural basis for allene oxide cyclization in moss | | Descriptor: | (9Z)-11-{(2R,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, (9Z)-11-{(2S,3S)-3-[(2Z)-pent-2-en-1-yl]oxiran-2-yl}undec-9-enoic acid, Allene oxide cyclase, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of Physcomitrella patens AOC1 and AOC2: Insights into the Enzyme Mechanism and Differences in Substrate Specificity.
Plant Physiol., 160, 2012
|
|
4ZLE
 
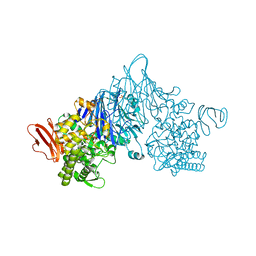 | | Cellobionic acid phosphorylase - ligand free structure | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
6UXY
 
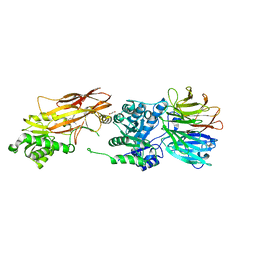 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 8 | | Descriptor: | (5R)-2-amino-5-(2-cyclohexylethyl)-3-methyl-5-phenyl-3,5-dihydro-4H-imidazol-4-one, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
3BFE
 
 | |
6V16
 
 | | Crystal structure of the bromodomain of human BRD7 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6IH7
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - 3',3'-cGAMP bound form | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, cyclic di nucleotide phoshodiesterase | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication.
Biochem.J., 476, 2019
|
|
7VB6
 
 | | Crystal structure of hydroxynitrile lyase from Linum usitatissium complexed with (R)-2-hydroxy-2-methylbutanenitrile | | Descriptor: | (2R)-2-methyl-2-oxidanyl-butanenitrile, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
6V3K
 
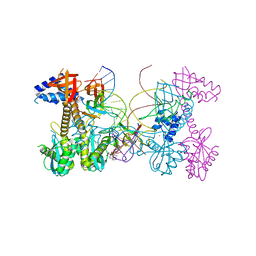 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ419 (compound 4c) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-(5-oxidanylpentyl)-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-11-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
2LD2
 
 | |
9MQB
 
 | | Vitamin K-dependent gamma-carboxylase with Osteocalcin (mutant) and vitamin K hydroquinone and calcium | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2025-01-02 | | Release date: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into the vitamin K-dependent gamma-carboxylation of osteocalcin.
Cell Res., 35, 2025
|
|
9NMP
 
 | | Structure of mouse RyR1 with simvastatin (Ca2+/CFF/ATP dataset; open pore) | | Descriptor: | (1S,3R,7S,8S,8aR)-8-{2-[(2R,4R)-4-hydroxy-6-oxooxan-2-yl]ethyl}-3,7-dimethyl-1,2,3,7,8,8a-hexahydronaphthalen-1-yl 2,2-dimethylbutanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2025-03-04 | | Release date: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for statin-induced skeletal muscle weakness
To Be Published
|
|
6V94
 
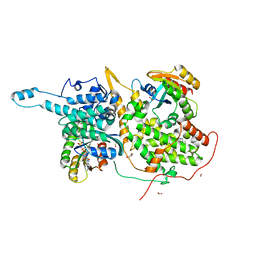 | |
9MQE
 
 | | Vitamin K-dependent gamma-carboxylase with Osteocalcin and vitamin K hydroquinone | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Liu, B, Cao, Q. | | Deposit date: | 2025-01-02 | | Release date: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into the vitamin K-dependent gamma-carboxylation of osteocalcin.
Cell Res., 35, 2025
|
|
8GAS
 
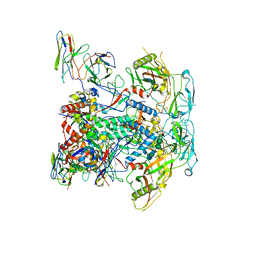 | | vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-23 | | Release date: | 2023-04-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
