4Q2Z
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE356, from a non-human primate | | Descriptor: | Heavy chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, Light chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody | | Authors: | Navis, M, Tran, K, Bale, S, Phad, G, Guenaga, J, Wilson, R, Soldemo, M, McKee, K, Sundling, C, Mascola, J, Li, Y, Wyatt, R.T, Hedestam, G.B.K. | | Deposit date: | 2014-04-10 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Plos Pathog., 10, 2014
|
|
4Q3I
 
 | | Structure of the OsSERK2 leucine rich repeat extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, OsSERK2 D128N | | Authors: | McAndrew, R.P, Pruitt, R.N, Kamita, S.G, Pereira, J.H, Majumder, D, Hammock, B.D, Adams, P.D, Ronald, P.C. | | Deposit date: | 2014-04-11 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the OsSERK2 leucine-rich repeat extracellular domain.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5SX6
 
 | | Crystal structure of the catalase-peroxidase KatG of B. pseudomallei at pH 6.5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles for Arg426 and Trp111 in the modulation of NADH oxidase activity of the catalase-peroxidase KatG from Burkholderia pseudomallei inferred from pH-induced structural changes.
Biochemistry, 45, 2006
|
|
5SX7
 
 | | Crystal structure of catalase-peroxidase KatG of B. pseudomallei at pH 8.5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Roles for Arg426 and Trp111 in the modulation of NADH oxidase activity of the catalase-peroxidase KatG from Burkholderia pseudomallei inferred from pH-induced structural changes.
Biochemistry, 45, 2006
|
|
5SYX
 
 | |
4Q5N
 
 | |
4Q6A
 
 | |
4Q7B
 
 | |
4LM7
 
 | |
1UTX
 
 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
5T5V
 
 | | LIPOXYGENASE-1 (SOYBEAN) AT 293K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
4Q9C
 
 | | IgNAR antibody domain C3 | | Descriptor: | CHLORIDE ION, Novel antigen receptor, SODIUM ION, ... | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZU0
 
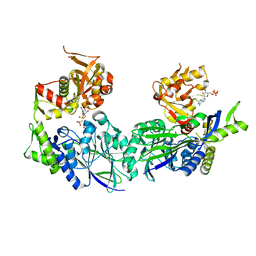 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | NAD NUCLEOTIDASE, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-13 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
1Z3J
 
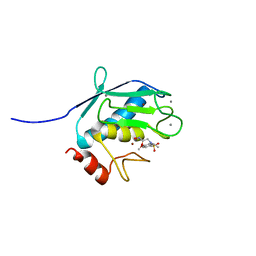 | | Solution Structure of MMP12 in the presence of N-isobutyl-N-4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.M, Luchinat, C, Mangani, S, Terni, B, Turano, P. | | Deposit date: | 2005-03-13 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5SW4
 
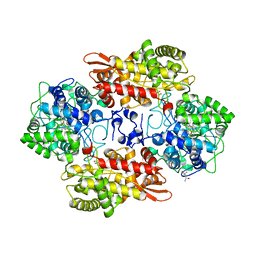 | | Crystal structure of native catalase-peroxidase KatG at pH8.0 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-08 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A molecular switch and electronic circuit modulate catalase activity in catalase-peroxidases.
EMBO Rep., 6, 2005
|
|
1US0
 
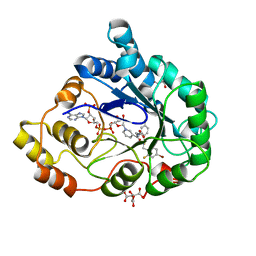 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | Authors: | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | Deposit date: | 2003-11-16 | | Release date: | 2004-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.66 Å) | | Cite: | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
4LML
 
 | | GLIC double mutant I9'A T25'A | | Descriptor: | Proton-gated ion channel | | Authors: | Grosman, C, Gonzalez-Gutierrez, G. | | Deposit date: | 2013-07-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZVH
 
 | | Methylaspartate ammonia lyase from Clostridium tetanomorphum mutant Q73A | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Raj, H, Szymanski, W, de Villiers, J, Rozeboom, H.J, Veetil, V.P, Reis, C.R, de Villiers, M, de Wildeman, S, Dekker, F.J, Quax, W.J, Thunnissen, A.M.W.H, Feringa, B.L, Janssen, D.B, Poelarends, G.J. | | Deposit date: | 2011-07-25 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Nat.Chem., 4, 2012
|
|
4QEW
 
 | | Crystal structure of BRD2(BD2) mutant with ligand ET bound (METHYL (2R)- 2-[(4S)-6-(4-CHLOROPHENYL)-8-METHOXY-1-METHYL-4H-[1,2,4]TRIAZOLO[4,3-A][1, 4]BENZODIAZEPIN-4-YL]BUTANOATE) | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Tallant, C, Baud, M, Lin-Shiao, E, Chirgadze, D.Y, Ciulli, A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes.
Science, 346, 2014
|
|
1UOE
 
 | | Crystal structure of the dihydroxyacetone kinase from E. coli in complex with glyceraldehyde | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCEROL, SULFATE ION | | Authors: | Siebold, C, Garcia-Alles, L.F, Luthi-Nyffeler, T, Flukiger-Bruhwiler, K, Burgi, H.-B, Baumann, U, Erni, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoenolpyruvate- and ATP-Dependent Dihydroxyacetone Kinases: Covalent Substrate-Binding and Kinetic Mechanism
Biochemistry, 43, 2004
|
|
3ZLM
 
 | |
1V0A
 
 | | Family 11 Carbohydrate-Binding Module of cellulosomal cellulase Lic26A-Cel5E of Clostridium thermocellum | | Descriptor: | CALCIUM ION, ENDOGLUCANASE H, SULFATE ION | | Authors: | Carvalho, A.L, Romao, M.J, Goyal, A, Prates, J.A.M, Pires, V.M.R, Ferreira, L.M.A, Bolam, D.N, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Family 11 Carbohydrate-Binding Module of Clostridium Thermocellum Lic26A-Cel5E Accomodates Beta-1,4- and Beta-1,3-1,4-Mixed Linked Glucans at a Single Binding Site
J.Biol.Chem., 279, 2004
|
|
3ZVZ
 
 | | PHD finger of human UHRF1 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE UHRF1, ZINC ION | | Authors: | Lallous, N, Birck, C, Mc Ewen, A.G, Legrand, P, Samama, J.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | The Phd Finger of Human Uhrf1 Reveals a New Subgroup of Unmethylated Histone H3 Tail Readers.
Plos One, 6, 2011
|
|
1YXS
 
 | | Crystal Structure of Kinase Pim1 with P123M mutation | | Descriptor: | IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of proto-oncogene kinase Pim1: a target of aberrant somatic hypermutations in diffuse large cell lymphoma.
J.Mol.Biol., 348, 2005
|
|
1UOD
 
 | | Crystal structure of the dihydroxyacetone kinase from E. coli in complex with dihydroxyacetone-phosphate | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCERALDEHYDE-3-PHOSPHATE, SULFATE ION | | Authors: | Siebold, C, Garcia-Alles, L.F, Luthi-Nyffeler, T, Flukiger-Bruhwiler, K, Burgi, H.-B, Baumann, U, Erni, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-09-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoenolpyruvate- and ATP-Dependent Dihydroxyacetone Kinases: Covalent Substrate-Binding and Kinetic Mechanism
Biochemistry, 43, 2004
|
|
