1E3D
 
 | | [NiFe] Hydrogenase from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), FE3-S4 CLUSTER, ... | | Authors: | Matias, P.M, Soares, C.M, Saraiva, L.M, Coelho, R, Morais, J, Le Gall, J, Carrondo, M.A. | | Deposit date: | 2000-06-14 | | Release date: | 2001-06-14 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | [Nife] Hydrogenase from Desulfovibrio Desulfuricans Atcc 27774: Gene Sequencing, Three-Dimensional Structure Determination and Refinement at 1.8 A and Modelling Studies of its Interaction with the Tetrahaem Cytochrome C3.
J.Biol.Inorg.Chem., 6, 2001
|
|
3URZ
 
 | |
3V7O
 
 | |
4FV2
 
 | | Crystal Structure of the ERK2 complexed with EK5 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-chlorophenyl)-1H-pyrazol-5-yl]-N-(2,3-dihydro-1-benzofuran-5-ylmethyl)-1H-pyrrole-2-carboxamide, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the ERK2 complexed with EK5
TO BE PUBLISHED
|
|
3MPX
 
 | | Crystal structure of the DH and PH-1 domains of human FGD5 | | Descriptor: | FYVE, RhoGEF and PH domain-containing protein 5, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Nedyalkova, L, Tong, Y, Tempel, W, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the DH and PH-1 domains of human FGD5
TO BE PUBLISHED
|
|
6ZT0
 
 | | Crystal structure of the Eiger TNF domain/Grindelwald extracellular domain complex | | Descriptor: | 1,2-ETHANEDIOL, Protein eiger, Protein grindelwald | | Authors: | Palmerini, V, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions.
Nat Commun, 12, 2021
|
|
9G7Q
 
 | | Structure of the StayRose dimer | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, StayRose | | Authors: | Crow, A. | | Deposit date: | 2024-07-22 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | StayRose: a photostable StayGold derivative red-shifted by genetic code expansion
Biorxiv, 2024
|
|
7A9A
 
 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
9FXS
 
 | |
7A6W
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 33-(Z) | | Descriptor: | (2S,9S,12R,20Z)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-28,31-dimethoxy-11,18,23,26-tetraoxa-4-azatetracyclo[25.2.2.113,17.04,9]dotriaconta-1(29),13(32),14,16,20,27,30-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AEM
 
 | | Studies Towards a Reversible EGFR C797S Triple Mutant Inhibitor Series | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Abstract 4451: Evaluation of the therapeutic potential of phosphine oxide pyrazole inhibitors in tumors harboring EGFR C797S mutation
Cancer Res., 79, 2019
|
|
1MQH
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Bromo-Willardiine at 1.8 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
7A50
 
 | | Crystal structure of the APH coiled-coil in complex with nanobody Nb26 | | Descriptor: | 1,2-ETHANEDIOL, Coiled-coil APH, Nanobody Nb26 | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1EAS
 
 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 3. DESIGN, SYNTHESIS, X-RAY CRYSTALLOGRAPHIC ANALYSIS, AND STRUCTURE-ACTIVITY RELATIONSHIPS FOR A SERIES OF ORALLY ACTIVE 3-AMINO-6-PHENYLPYRIDIN-2-ONE TRIFLUOROMETHYL KETONES | | Descriptor: | 3-[[(METHYLAMINO)SULFONYL]AMINO]-2-OXO-6-PHENYL-N-[3,3,3-TRIFLUORO-1-(1-METHYLETHYL)-2-OXOPHENYL]-1(2H)-PYRIDINE ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 3. Design, synthesis, X-ray crystallographic analysis, and structure-activity relationships for a series of orally active 3-amino-6-phenylpyridin-2-one trifluoromethyl ketones.
J.Med.Chem., 37, 1994
|
|
4GHI
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains in complex with a benzoxadiazole antagonist | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Scheuermann, T.H, Key, J, Tambar, U.K, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2012-08-07 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric inhibition of hypoxia inducible factor-2 with small molecules.
Nat.Chem.Biol., 9, 2013
|
|
2OQV
 
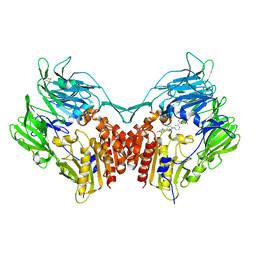 | | Human Dipeptidyl Peptidase IV (DPP4) with piperidine-constrained phenethylamine | | Descriptor: | (3R,4R)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-3-AMINE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D. | | Deposit date: | 2007-02-01 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
4FTG
 
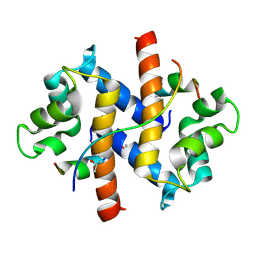 | |
6SI5
 
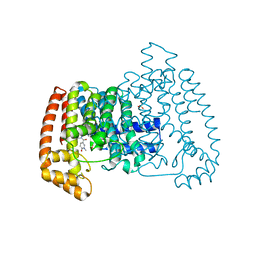 | | T. cruzi FPPS in complex with 1-methyl-5-(4,5,6,7-tetrahydrothieno[3,2-c]pyridine-5-carbonyl)pyridin-2(1H)-one | | Descriptor: | 5-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylcarbonyl)-1-methyl-pyridin-2-one, Farnesyl diphosphate synthase, SULFATE ION | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Cornaciu, I, Clavel, D, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
6ZOE
 
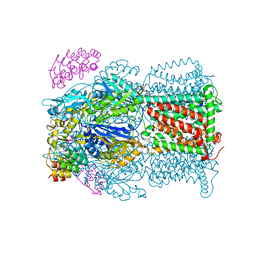 | | AcrB-F563A symmetric T protomer | | Descriptor: | 1,2-ETHANEDIOL, DARPIN, DECANE, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
6ZOA
 
 | | Partially induced AcrB T protomer and DDM binding to the TM8/PC2 pathway of AcrB L2 protomer | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DARPIN, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
7A74
 
 | | Structure of G132N BlaC from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two beta-Lactamase Variants with Reduced Clavulanic Acid Inhibition Display Different Millisecond Dynamics.
Antimicrob.Agents Chemother., 65, 2021
|
|
1ESK
 
 | | SOLUTION STRUCTURE OF NCP7 FROM HIV-1 | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Morellet, N, Demene, H, Teilleux, V, Huynh-Dinh, T, de Rocquigny, H, Fournie-Zaluski, M.-C, Roques, B.P. | | Deposit date: | 2000-04-10 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of (12-53)NCp7 of HIV-1
To be Published
|
|
4FTV
 
 | |
3MWW
 
 | | Crystal structure of HCV NS5B polymerase | | Descriptor: | 1-[2-(4-carboxypiperidin-1-yl)-2-oxoethyl]-3-cyclohexyl-2-furan-3-yl-1H-indole-6-carboxylic acid, Genome polyprotein, SULFATE ION | | Authors: | Coulombe, R. | | Deposit date: | 2010-05-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
9FXX
 
 | |
