7T7D
 
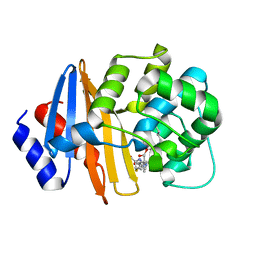 | | MA-1-206-OXA-23 30s complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
6UY9
 
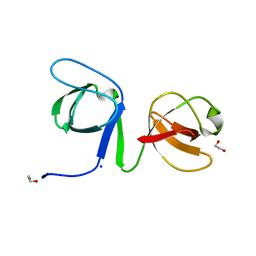 | |
8EL0
 
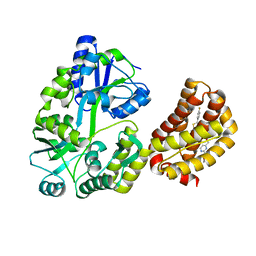 | | Structure of MBP-Mcl-1 in complex with a macrocyclic compound | | Descriptor: | (7R,20P)-18-chloro-1-(4-fluorophenyl)-10-{[(2M)-2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy}-19-methyl-15-[2-(4-methylpiperazin-1-yl)ethyl]-7,8,15,16-tetrahydro-14H-17,20-etheno-9,13-(metheno)-6-oxa-2-thia-3,5,15-triazacyclooctadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
6FMY
 
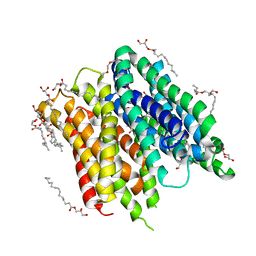 | | IMISX-EP of S-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
8EKX
 
 | | Structure of MBP-Mcl-1 in complex with MIK665 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
8TUA
 
 | | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)
Elife, 2024
|
|
6VLF
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
8F9Z
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-14204, an HIV-1 gp120 antagonist | | Descriptor: | (5M)-N-{(1S)-2-amino-1-[5-(hydroxymethyl)-1,3-thiazol-2-yl]ethyl}-5-[5-(trifluoromethyl)pyridin-2-yl]-1H-pyrrole-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Antiviral Activity and Crystal Structures of HIV-1 gp120 Antagonists.
Int J Mol Sci, 23, 2022
|
|
7NSN
 
 | | Multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis: mannoimidazole complex | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Kolaczkowski, B.M, Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S, Moeler, M.S, Meyer, A.S, Westh, P, Jensen, K, Krogh, K.B.R.M. | | Deposit date: | 2021-03-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and functional characterization of a multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6W04
 
 | |
8OUB
 
 | |
6VX2
 
 | |
6W2G
 
 | | Crystal Structure of Y188G Variant of the Internal UBA Domain of HHR23A in Monoclinic Unit Cell | | Descriptor: | 1,2-ETHANEDIOL, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
6YUL
 
 | | CK2 alpha bound to Macrocycle | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
7V3Z
 
 | | Structure of cannabinoid receptor type 1(CB1) | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, CHOLESTEROL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liu, Z.J, Shen, L, Hua, T, Yao, D.Q, Wu, L.J. | | Deposit date: | 2021-08-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | A Genetically Encoded F-19 NMR Probe Reveals the Allosteric Modulation Mechanism of Cannabinoid Receptor 1.
J.Am.Chem.Soc., 143, 2021
|
|
8ZP0
 
 | | Cryo-EM structure of YF23694-bound porcine bc1 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1-[2-[(4,6-dimethyl-1,3-benzothiazol-2-yl)sulfanylmethyl]-3-methyl-phenyl]-4-methyl-1,2,3,4-tetrazol-5-one, CARDIOLIPIN, ... | | Authors: | Wang, Y.X, Sun, J.Y, Cui, G.R, Yang, G.F. | | Deposit date: | 2024-05-29 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8ZOW
 
 | | Cryo-EM structure of Metyltetraprole-bound porcine bc1 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1-[2-[[1-(4-chlorophenyl)pyrazol-3-yl]oxymethyl]-3-methyl-phenyl]-4-methyl-1,2,3,4-tetrazol-5-one, CARDIOLIPIN, ... | | Authors: | Wang, Y.X, Sun, J.Y, Cui, G.R, Yang, G.F. | | Deposit date: | 2024-05-29 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
6X47
 
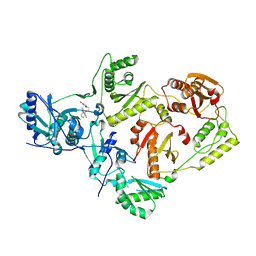 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ649), a Non-nucleoside Inhibitor | | Descriptor: | 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.767 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6X49
 
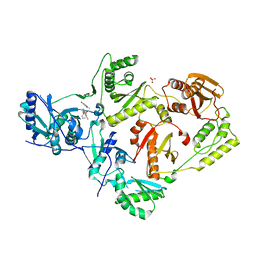 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ649), a Non-nucleoside Inhibitor | | Descriptor: | 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6VZH
 
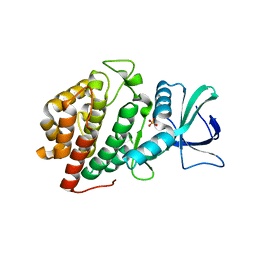 | | Structure of Human Vaccinia-related Kinase 1 (VRK1) Bound to LDSM311 | | Descriptor: | (7~{R})-2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7-dimethyl-8-prop-2-ynyl-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | dos Reis, C.V, Dutra, L.A, Gama, F, Ferreira, M, Mascarello, A, Azevedo, H, Guimaraes, C, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Human Vaccinia-related Kinase 1 (VRK1) Bound to LDSM311
To be Published
|
|
8ZYT
 
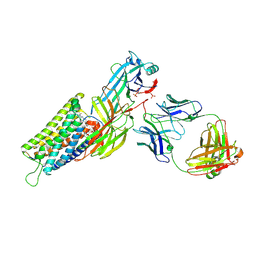 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin1 and SBI-553 (complex 3) | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Sun, D, Li, X, Yuan, Q, Yin, W, Xu, H.E, Tian, C. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
8ZYY
 
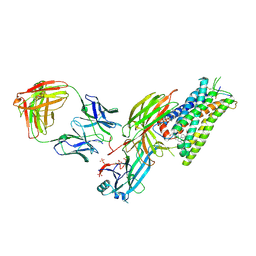 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin1 and SBI-553 (complex 2) | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Sun, D, Li, X, Yuan, Q, Yin, W, Xu, H.E, Tian, C. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
6X4A
 
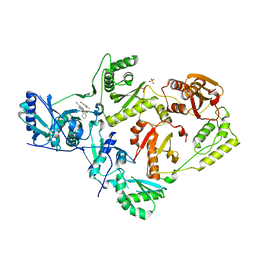 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | Descriptor: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
8UA5
 
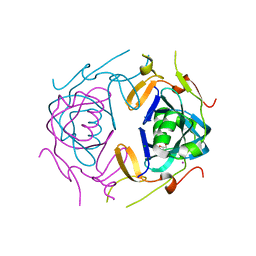 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (A636-Q776) | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 92, 2024
|
|
8G8W
 
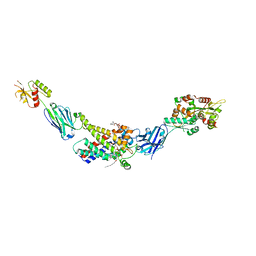 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
