2I2H
 
 | | NMR structure of TPC3 in TFE | | Descriptor: | signaling peptide TCP3 | | Authors: | Syvitski, R.T, Jakeman, D.L, Li, Y. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Analysis of Quorum-Sensing Signaling Peptides from Streptococcus mutans.
J.Bacteriol., 189, 2007
|
|
2I2I
 
 | | Crystal structure of the DB293-D(CGCGAATTCGCG)2 complex. | | Descriptor: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-FURYL)-1H-BENZIMIDAZOLE-5-CARBOXIMIDAMIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Lee, M.P.H, Neidle, S. | | Deposit date: | 2006-08-16 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the DB293-D(CGCGAATTCGCG)2 complex.
To be Published
|
|
2I2J
 
 | | NMR structure of UA159sp in TFE | | Descriptor: | Competence stimulating peptide | | Authors: | Syvitski, R.T, Jakeman, D.L, Li, Y. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2020-03-04 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Analysis of Quorum-Sensing Signaling Peptides from Streptococcus mutans.
J.Bacteriol., 189, 2007
|
|
2I2L
 
 | | X-ray Crystal Structure of Protein yopX from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR411. | | Descriptor: | YopX protein | | Authors: | Vorobiev, S.M, Zhou, W, Seetharaman, J, Forouhar, F, Kuzin, A.A, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-16 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein yopX from Bacillus subtilis
To be Published
|
|
2I2O
 
 | | Crystal Structure of an eIF4G-like Protein from Danio rerio | | Descriptor: | NICKEL (II) ION, eIF4G-like protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-16 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of an eIF4G-like protein from Danio rerio.
Proteins, 78, 2010
|
|
2I2Q
 
 | | Fission Yeast cofilin | | Descriptor: | 1,2-ETHANEDIOL, Cofilin, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Andrianantoandro, E, Pollard, T.D. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Mechanism of Actin Filament Turnover by Severing and Nucleation at Different Concentrations of ADF/Cofilin.
Mol.Cell, 24, 2006
|
|
2I2R
 
 | | Crystal structure of the KChIP1/Kv4.3 T1 complex | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Findeisen, F, Pioletti, M, Minor Jr, D.L. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer
Nat.Struct.Mol.Biol., 13, 2006
|
|
2I2S
 
 | | Crystal Structure of the porcine CRW-8 rotavirus VP8* carbohydrate-recognising domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, ... | | Authors: | Blanchard, H. | | Deposit date: | 2006-08-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into Host Cell Carbohydrate-recognition by Human and Porcine Rotavirus from Crystal Structures of the Virion Spike Associated Carbohydrate-binding Domain (VP8*)
J.Mol.Biol., 367, 2007
|
|
2I2W
 
 | | Crystal Structure of Escherichia Coli Phosphoheptose Isomerase | | Descriptor: | GLYCEROL, Phosphoheptose isomerase | | Authors: | DeLeon, G, Blakely, K, Zhang, K, Wright, G, Junop, M. | | Deposit date: | 2006-08-17 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2I2X
 
 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | Authors: | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I2Y
 
 | | Solution structure of the RRM of SRp20 bound to the RNA CAUC | | Descriptor: | (5'-R(*CP*AP*UP*C)-3'), Fusion protein consists of immunoglobulin G-Binding Protein G and Splicing factor, arginine/serine-rich 3 | | Authors: | Hargous, Y.F, Allain, F.H. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.
Embo J., 25, 2006
|
|
2I2Z
 
 | | Human serum albumin complexed with myristate and aspirin | | Descriptor: | 2-HYDROXYBENZOIC ACID, MYRISTIC ACID, Serum albumin | | Authors: | Yang, F, Bian, C, Zhu, L, Zhao, G, Huang, Z, Huang, M. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of human serum albumin on drug metabolism: Structural evidence of esterase activity of human serum albumin
J.Struct.Biol., 157, 2007
|
|
2I30
 
 | | Human serum albumin complexed with myristate and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, MYRISTIC ACID, Serum albumin | | Authors: | Yang, F, Bian, C, Zhu, L, Zhao, G, Huang, Z, Huang, M. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Effect of human serum albumin on drug metabolism: Structural evidence of esterase activity of human serum albumin
J.Struct.Biol., 157, 2007
|
|
2I32
 
 | |
2I33
 
 | |
2I34
 
 | |
2I35
 
 | | Crystal structure of rhombohedral crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I36
 
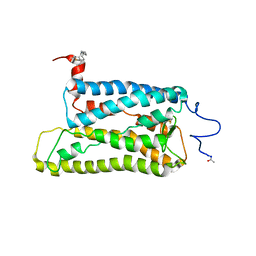 | | Crystal structure of trigonal crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, Rhodopsin, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I37
 
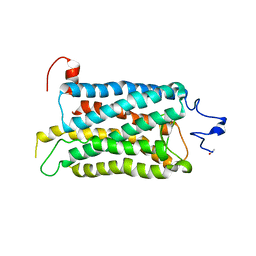 | | Crystal structure of a photoactivated rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rhodopsin, ... | | Authors: | Lodowski, D.T, Stenkamp, R.E, Salom, D, Le Trong, I, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I38
 
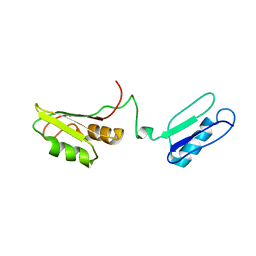 | | Solution structure of the RRM of SRp20 | | Descriptor: | Fusion protein consists of immunoglobulin G-Binding Protein G and Splicing factor, arginine/serine-rich 3 | | Authors: | Hargous, Y.F, Allain, F.H.T. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.
Embo J., 25, 2006
|
|
2I39
 
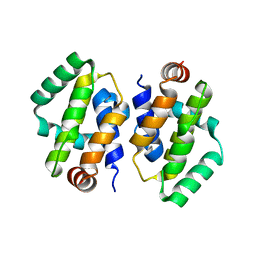 | | Crystal structure of Vaccinia virus N1L protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein N1 | | Authors: | Aoyagi, M, Aleshin, A.E, Stec, B, Liddington, R.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein.
Protein Sci., 16, 2007
|
|
2I3A
 
 | | Crystal structure of N-Acetyl-gamma-Glutamyl-Phosphate Reductase (Rv1652) from Mycobacterium tuberculosis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Moraidin, F, James, M.N.G, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of N-acetyl-gamma-glutamyl-phosphate Reductase from Mycobacterium tuberculosis in Complex with NADP(+).
J.Mol.Biol., 367, 2007
|
|
2I3B
 
 | |
2I3C
 
 | | Crystal Structure of an Aspartoacylase from Homo Sapiens | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I3D
 
 | | Crystal Structure of Protein of Unknown Function ATU1826, a Putative Alpha/Beta Hydrolase from Agrobacterium tumefaciens | | Descriptor: | CHLORIDE ION, Hypothetical protein Atu1826, MAGNESIUM ION | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-17 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein Atu1826, a putative alpha/beta hydrolase from Agrobacterium tumefaciens.
To be Published
|
|
