4KG0
 
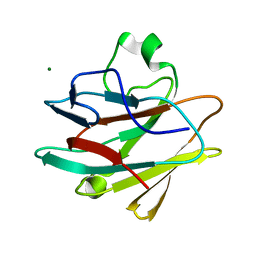 | | Crystal structure of the drosophila melanogaster neuralized-nhr1 domain | | Descriptor: | MAGNESIUM ION, Protein neuralized | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2013-04-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure, biochemical and biophysical characterisation of NHR1 domain of E3 Ubiquitin ligase neutralized
Advances in Enzyme Research, 1, 2013
|
|
7RL3
 
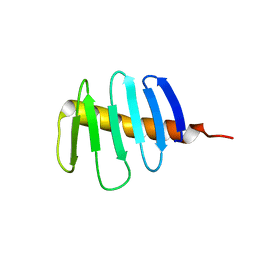 | | Structure of Drosophila melanogaster Plk4 PB3 | | Descriptor: | Serine/threonine-protein kinase PLK4 | | Authors: | Slep, K.C, Slevin, L.K. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Polo-like kinase 4 homodimerization and condensate formation regulate its own protein levels but are not required for centriole assembly.
Mol.Biol.Cell, 34, 2023
|
|
5LSW
 
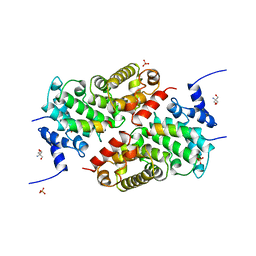 | | A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell differentiation protein RCD1 homolog, LD12033p, ... | | Authors: | Sgromo, A, Raisch, T, Bawankar, P, Bhandari, D, Chen, Y, Kuzuoglu-Ozturk, D, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-09-05 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin.
Nat Commun, 8, 2017
|
|
1FTZ
 
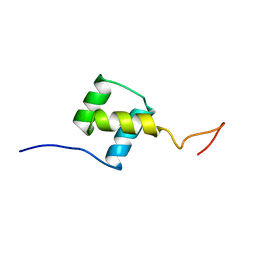 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE FUSHI TARAZU HOMEODOMAIN FROM DROSOPHILA AND COMPARISON WITH THE ANTENNAPEDIA HOMEODOMAIN | | Descriptor: | FUSHI TARAZU PROTEIN | | Authors: | Qian, Y.Q, Furukubo-Tokunaga, K, Resendez-Perez, D, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain.
J.Mol.Biol., 238, 1994
|
|
2NCD
 
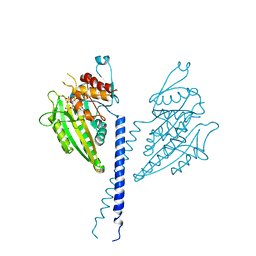 | | NCD (NON-CLARET DISJUNCTIONAL) DIMER FROM D. MELANOGASTER | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROTEIN (Kinesin motor NCD), SULFATE ION | | Authors: | Sablin, E.P, Case, R.B, Dai, S.C, Hart, C.L, Ruby, A, Vale, R.D, Fletterick, R.J. | | Deposit date: | 1999-06-04 | | Release date: | 1999-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direction determination in the minus-end-directed kinesin motor ncd.
Nature, 395, 1998
|
|
3KNF
 
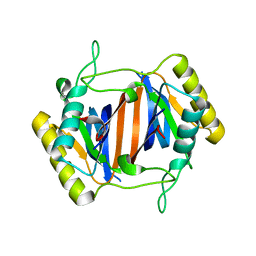 | |
6S9R
 
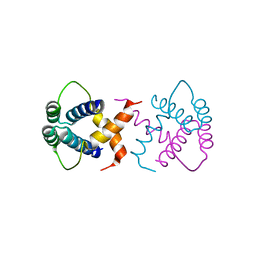 | | Crystal structure of SSDP from D. melanogaster | | Descriptor: | Sequence-specific single-stranded DNA-binding protein, isoform A | | Authors: | Renko, M, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3KO3
 
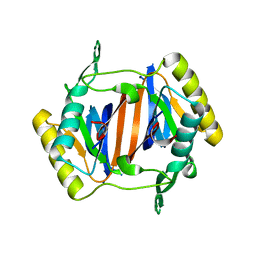 | |
3KOD
 
 | |
3KO7
 
 | |
3KO9
 
 | |
8EW6
 
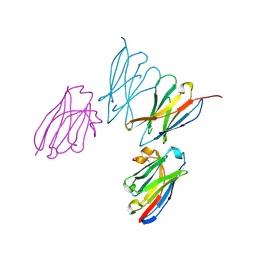 | | Anti-human CD8 VHH complex with CD8 alpha | | Descriptor: | Anti-CD8 alpha VHH, T-cell surface glycoprotein CD8 alpha chain | | Authors: | Kiefer, J.R, Williams, S, Davies, C.W, Koerber, J.T, Sriraman, S.K, Yin, Y.P. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Development of an 18 F-labeled anti-human CD8 VHH for same-day immunoPET imaging.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
3KO5
 
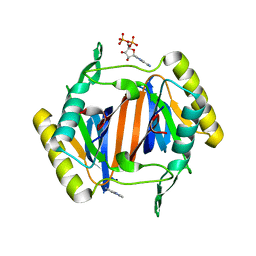 | |
5GKB
 
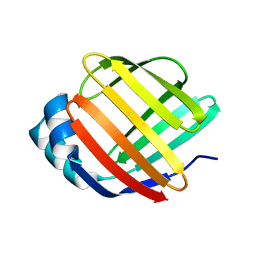 | | Crystal Structure of Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster without citrate inside | | Descriptor: | Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-07-04 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
5GGE
 
 | | Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster | | Descriptor: | CITRIC ACID, Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
8EDU
 
 | | Mycobacteriophage Muddy capsid | | Descriptor: | Capsid | | Authors: | Freeman, K.G, White, S.J, Huet, A, Conway, J.F. | | Deposit date: | 2022-09-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
3LRS
 
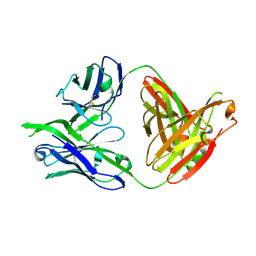 | | Structure of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PG-16 Heavy Chain Fab, PG-16 Light Chain Fab | | Authors: | Pancera, M, Kwong, P.D. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
8EEC
 
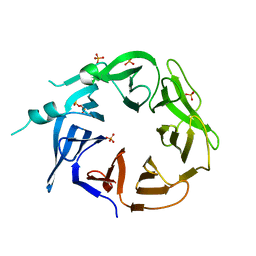 | | Crystal structure of HPK1 citron-homology domain | | Descriptor: | Isoform 2 of Mitogen-activated protein kinase kinase kinase kinase 1, PHOSPHATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2022-09-06 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | HPK1 Citron Homology Domain Serves as a Scaffold to Promote Phosphorylation of SLP76
To Be Published
|
|
4NV0
 
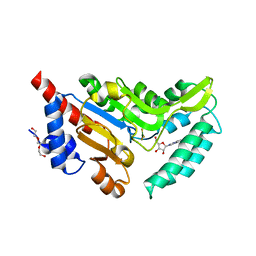 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to 7-methylguanosine | | Descriptor: | 7-METHYLGUANOSINE, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
8G0L
 
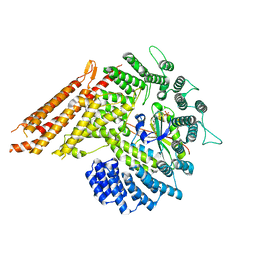 | |
8FG8
 
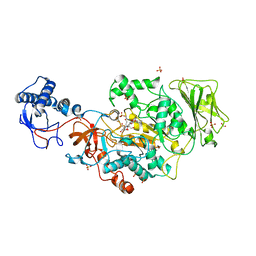 | | Catalytic domain of GtfB in complex with inhibitor 2-[(2,4,5-Trihydroxyphenyl)methylidene]-1-benzofuran-3-one | | Descriptor: | (2Z)-2-[(2,4,5-trihydroxyphenyl)methylidene]-1-benzofuran-3(2H)-one, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C, Velu, S. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Hydrogel-Encapsulated Biofilm Inhibitors Abrogate the Cariogenic Activity of Streptococcus mutans .
J.Med.Chem., 66, 2023
|
|
3MFK
 
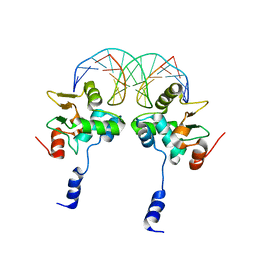 | |
3MF5
 
 | |
7VKK
 
 | | Crystal structure of D. melanogaster SAMTOR V66W/E67P mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine sensor upstream of mTORC1, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of S -adenosylmethionine sensing by SAMTOR in mTORC1 signaling.
Sci Adv, 8, 2022
|
|
7VKR
 
 | |
