2HQ8
 
 | | Crystal structure of coelenterazine-binding protein from renilla muelleri in the ca loaded apo form | | Descriptor: | CALCIUM ION, Coelenterazine-binding protein ca-bound apo form | | Authors: | Stepanyuk, G, Liu, Z.J, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of coelenterazine-binding protein from Renilla muelleri at 1.7 A: why it is not a calcium-regulated photoprotein.
PHOTOCHEM.PHOTOBIOL.SCI., 7, 2008
|
|
2HQ9
 
 | |
2HQA
 
 | | Crystal structure of the catalytic alpha subunit of E. Coli replicative DNA polymerase III | | Descriptor: | DNA polymerase III alpha subunit, PHOSPHATE ION | | Authors: | Lamers, M.H, Georgescu, R.E, Lee, S.G, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HQB
 
 | |
2HQC
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQD
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQE
 
 | | Crystal structure of human P100 Tudor domain: Large fragment | | Descriptor: | P100 Co-activator tudor domain | | Authors: | Shah, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Liu, Z.J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a large fragment of the Human P100 Tudor Domain
To be Published
|
|
2HQF
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQG
 
 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | Descriptor: | Acriflavine resistance protein B | | Authors: | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | Deposit date: | 2006-07-18 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
2HQH
 
 | | Crystal structure of p150Glued and CLIP-170 | | Descriptor: | Dynactin-1, Restin, ZINC ION | | Authors: | Hayashi, I, Ikura, M. | | Deposit date: | 2006-07-18 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CLIP170 autoinhibition mimics intermolecular interactions with p150Glued or EB1.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2HQI
 
 | | NMR SOLUTION STRUCTURE OF THE OXIDIZED FORM OF MERP, 14 STRUCTURES | | Descriptor: | MERCURIC TRANSPORT PROTEIN | | Authors: | Qian, H, Sahlman, L, Eriksson, P.O, Hambreus, C, Edlund, U, Sethson, I. | | Deposit date: | 1998-03-31 | | Release date: | 1998-11-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the oxidized form of MerP, a mercuric ion binding protein involved in bacterial mercuric ion resistance.
Biochemistry, 37, 1998
|
|
2HQJ
 
 | |
2HQK
 
 | | Crystal structure of a monomeric cyan fluorescent protein derived from Clavularia | | Descriptor: | ACETATE ION, CHLORIDE ION, Cyan fluorescent chromoprotein, ... | | Authors: | Henderson, J.N, Campbell, R.E, Ai, H, Remington, S.J. | | Deposit date: | 2006-07-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging.
Biochem.J., 400, 2006
|
|
2HQL
 
 | |
2HQM
 
 | |
2HQN
 
 | |
2HQO
 
 | |
2HQP
 
 | |
2HQQ
 
 | | Crystal structure of human ketohexokinase complexed to different sugar molecules | | Descriptor: | Ketohexokinase, SULFATE ION | | Authors: | Trinh, C.H, Asipu, A, Bonthron, D.T, Phillips, S.E.V. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of alternatively spliced isoforms of human ketohexokinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2HQR
 
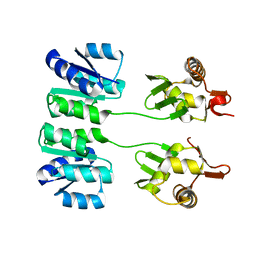 | |
2HQS
 
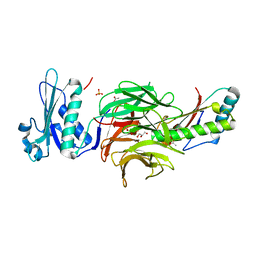 | | Crystal structure of TolB/Pal complex | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Grishkovskaya, I, Bonsor, D.A, Kleanthous, C, Dodson, E.J. | | Deposit date: | 2006-07-19 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mimicry enables competitive recruitment by a natively disordered protein.
J.Am.Chem.Soc., 129, 2007
|
|
2HQT
 
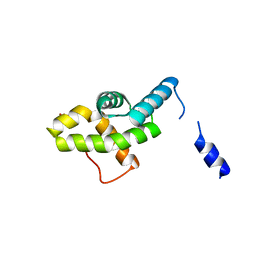 | |
2HQU
 
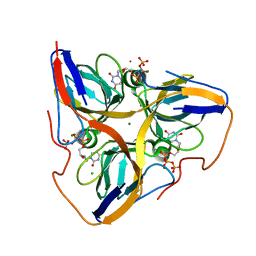 | | Human dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Varga, B, Vertessy, B.G. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase.
Febs Lett., 581, 2007
|
|
2HQV
 
 | | X-ray Crystal Structure of Protein AGR_C_4470 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR92. | | Descriptor: | AGR_C_4470p | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Zhao, L, Cunningham, K, Ma, L.C, Fang, Y, Xiao, R, Acton, T, Montelione, T.G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AGR_C_4470p from Agrobacterium tumefaciens.
Protein Sci., 16, 2007
|
|
2HQW
 
 | | Crystal Structure of Ca2+/Calmodulin bound to NMDA Receptor NR1C1 peptide | | Descriptor: | CALCIUM ION, Calmodulin, Glutamate NMDA receptor subunit zeta 1 | | Authors: | Akyol, Z, Gakhar, L, Sorensen, B.R, Hell, J.H, Shea, M.A. | | Deposit date: | 2006-07-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The NMDA Receptor NR1 C1 Region Bound to Calmodulin: Structural Insights into Functional Differences between Homologous Domains.
Structure, 15, 2007
|
|
