6SFI
 
 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SFJ
 
 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
2FNJ
 
 | |
6SFK
 
 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
7ZZO
 
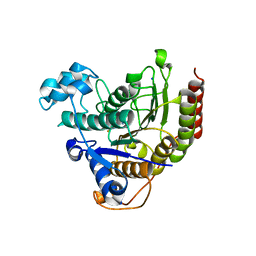 | | HDAC2 in complex with an inhibitor | | Descriptor: | 2-(cyclohexylazaniumyl)ethanesulfonate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZT
 
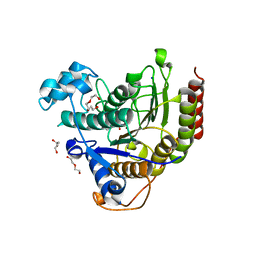 | | Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZP
 
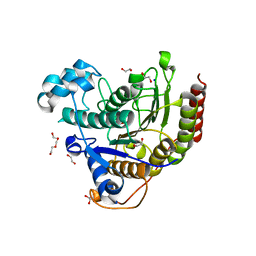 | | Structure of HDAC2 complexed with an inhibitory ligand | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZR
 
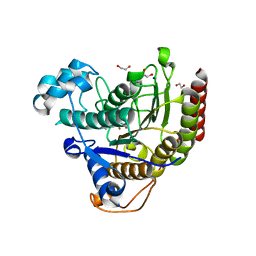 | | HDAC2 in complex with inhibitory ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
5LYS
 
 | | The crystal structure of 7SK 5'-hairpin - Gold derivative | | Descriptor: | 7SK RNA, GOLD ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
4DLJ
 
 | | Human p38 MAP kinase in complex with RL163 | | Descriptor: | 2-phenyl-N~4~-(2-phenylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Gruetter, C, Termathe, M. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fluorophore labeled kinase detects ligands that bind within the MAPK insert of p38alpha kinase.
Plos One, 7, 2012
|
|
7ZZW
 
 | | Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, CALCIUM ION, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZS
 
 | | HDAC2 complexed with an inhibitory ligand | | Descriptor: | (5~{S})-5-(4-chlorophenyl)pyrrolidin-2-one, 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
8A0B
 
 | | Inhibitor binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dihydroisoindol-2-yl-[(2R,4S)-4-phenylpyrrolidin-1-ium-2-yl]methanone, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZU
 
 | | Inhibitory Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R},4~{S})-4-phenylpyrrolidin-2-yl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
5MTX
 
 | | Dibenzooxepinone inhibitor 12b in complex with p38 MAPK | | Descriptor: | 3-[(3-benzamido-4-fluoranyl-phenyl)amino]-~{N}-(2-morpholin-4-ylethyl)-11-oxidanylidene-6~{H}-benzo[c][1]benzoxepine-9-carboxamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-01-11 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Novel Type I(1)/2 p38 alpha MAP Kinase Inhibitors with Excellent Selectivity, High Potency, and Prolonged Target Residence Time by Interfering with the R-Spine.
J. Med. Chem., 60, 2017
|
|
5MTY
 
 | | Dibenzosuberone inhibitor 8e in complex with p38 MAPK | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside, ~{N}-[2,4-bis(fluoranyl)-5-[[14-(2-hydroxyethylcarbamoyl)-2-oxidanylidene-6-tricyclo[9.4.0.0^{3,8}]pentadeca-1(15),3(8),4,6,11,13-hexaenyl]amino]phenyl]thiophene-2-carboxamide | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-01-11 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Novel Type I(1)/2 p38 alpha MAP Kinase Inhibitors with Excellent Selectivity, High Potency, and Prolonged Target Residence Time by Interfering with the R-Spine.
J. Med. Chem., 60, 2017
|
|
6PA7
 
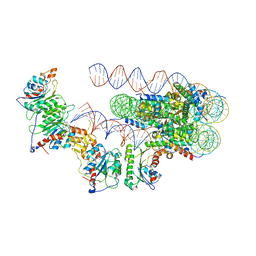 | | The cryo-EM structure of the human DNMT3A2-DNMT3B3 complex bound to nucleosome. | | Descriptor: | CHLORIDE ION, DNA (167-MER), DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Xu, T.H, Liu, M, Zhou, X.E, Liang, G, Zhao, G, Xu, H.E, Melcher, K, Jones, P.A. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Nature, 586, 2020
|
|
4MK5
 
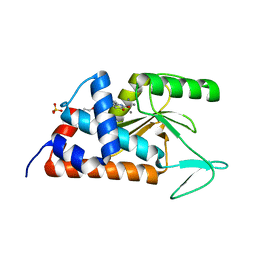 | | 6-(3-methoxyphenyl)pyridine-2,3-diol bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-(3-methoxyphenyl)pyridin-2(5H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
5LLI
 
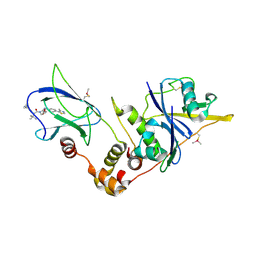 | | pVHL:EloB:EloC in complex with VH298 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-cyanocyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent and selective chemical probe of hypoxic signalling downstream of HIF-alpha hydroxylation via VHL inhibition.
Nat Commun, 7, 2016
|
|
6SN1
 
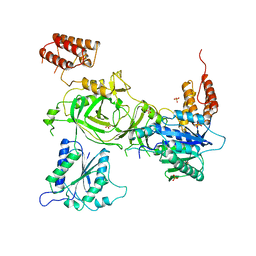 | | Crystal structure of the human INTS13-INTS14 complex | | Descriptor: | Integrator complex subunit 13, Integrator complex subunit 14, SULFATE ION | | Authors: | Jonas, S, Sabath, K, Staeubli, M.L. | | Deposit date: | 2019-08-23 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | INTS10-INTS13-INTS14 form a functional module of Integrator that binds nucleic acids and the cleavage module.
Nat Commun, 11, 2020
|
|
5LYV
 
 | | The crystal structure of 7SK 5'-hairpin - Osmium derivative | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
5LYU
 
 | | The native crystal structure of 7SK 5'-hairpin | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.C, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
6QZA
 
 | | HLA-DR1 with GMF Influenza PB1 Peptide | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Greenshields-Watson, A, Rizkallah, P.J. | | Deposit date: | 2019-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | CD4+T Cells Recognize Conserved Influenza A Epitopes through Shared Patterns of V-Gene Usage and Complementary Biochemical Features.
Cell Rep, 32, 2020
|
|
5T35
 
 | | The PROTAC MZ1 in complex with the second bromodomain of Brd4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Transcription elongation factor B polypeptide 1, ... | | Authors: | Gadd, M.S, Zengerle, M, Ciulli, A. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of PROTAC cooperative recognition for selective protein degradation.
Nat. Chem. Biol., 13, 2017
|
|
5VFU
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
