2FI4
 
 | | Crystal structure of a BPTI variant (Cys14->Ser) in complex with trypsin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural roles of the Cys14-Cys38 disulfide of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 382, 2008
|
|
6MC0
 
 | |
2J9Y
 
 | | Tryptophan Synthase Q114N mutant in complex with Compound II | | Descriptor: | (3E)-4-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}-2-IMINOBUT-3-ENOIC ACID, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Blumenstein, L, Domratcheva, T, Niks, D, Ngo, H, Seidel, R, Dunn, M.F, Schlichting, I. | | Deposit date: | 2006-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Betaq114N and Betat110V Mutations Reveal a Critically Important Role of the Substrate Alpha-Carboxylate Site in the Reaction Specificity of Tryptophan Synthase.
Biochemistry, 46, 2007
|
|
2J9Z
 
 | | Tryptophan Synthase T110 mutant complex | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Blumenstein, L, Domratcheva, T, Niks, D, Ngo, H, Seidel, R, Dunn, M.F, Schlichting, I. | | Deposit date: | 2006-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Betaq114N and Betat110V Mutations Reveal a Critically Important Role of the Substrate Alpha-Carboxylate Site in the Reaction Specificity of Tryptophan Synthase.
Biochemistry, 46, 2007
|
|
6M0F
 
 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) in substrate-free form | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Antibody fragment (9D5) Light chain, Antibody fragment (9D5) heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M3Z
 
 | | X-ray structure of a Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in milnacipran bound form | | Descriptor: | (1R,2S)-2-(aminomethyl)-N,N-diethyl-1-phenyl-cyclopropane-1-carboxamide, Antibody fragment 9D5 Light chain, Antibody fragment 9D5 heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-03-04 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M47
 
 | | X-ray structure of a Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in tramadol bound form | | Descriptor: | (1S,2S)-2-[(dimethylamino)methyl]-1-(3-methoxyphenyl)cyclohexan-1-ol, Antibody fragment 9D5 light chain, Antibody fragment Heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-03-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
4CBC
 
 | |
6KFN
 
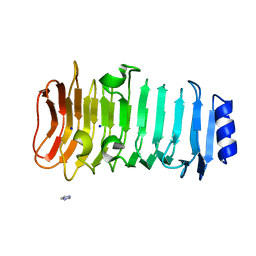 | | Crystal structure of alginate lyase from Paenibacillus sp. str. FPU-7 | | Descriptor: | IMIDAZOLE, SODIUM ION, alginate lyase | | Authors: | Itoh, T, Nakagawa, E, Yoda, M, Nakaichi, A, Hibi, T, Kimoto, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural and biochemical characterisation of a novel alginate lyase from Paenibacillus sp. str. FPU-7.
Sci Rep, 9, 2019
|
|
3ZYV
 
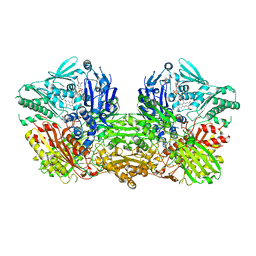 | | Crystal structure of the mouse liver Aldehyde Oxidase 3 (mAOX3) | | Descriptor: | AOX3, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Trincao, J, Coelho, C, Mahro, M, Rodrigues, D, Terao, M, Garattini, E, Leimkuehler, S, Romao, M.J. | | Deposit date: | 2011-08-27 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | The First Mammalian Aldehyde Oxidase Crystal Structure: Insights Into Substrate Specificity.
J.Biol.Chem., 287, 2012
|
|
3FIU
 
 | | Structure of NMN synthetase from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Sorci, L, Martynowski, D, Eyobo, Y, Osterman, A.L, Zhang, H. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nicotinamide mononucleotide synthetase is the key enzyme for an alternative route of NAD biosynthesis in Francisella tularensis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2YEQ
 
 | | Structure of PhoD | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALKALINE PHOSPHATASE D, ... | | Authors: | Lillington, J.E.D, Rodriguez, F, Roversi, P, Johnson, S.J, Berks, B, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Phosphodiesterase Phod Reveals an Iron and Calcium-Containing Active Site.
J.Biol.Chem., 289, 2014
|
|
4HMM
 
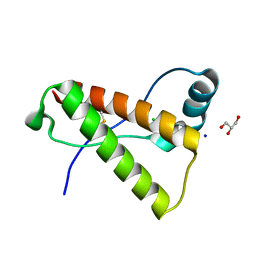 | | Crystal structure of mutant rabbit PRP 121-230 (S174N) | | Descriptor: | CHLORIDE ION, GLYCEROL, Major prion protein, ... | | Authors: | Sweeting, B, Chakrabartty, A, Pai, E.F. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N-Terminal Helix-Cap in alpha-Helix 2 Modulates beta-State Misfolding in Rabbit and Hamster Prion Proteins.
Plos One, 8, 2013
|
|
4FW6
 
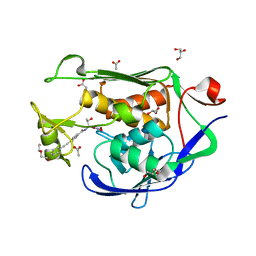 | |
2FI3
 
 | | Crystal structure of a BPTI variant (Cys14->Ser, Cys38->Ser) in complex with trypsin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Zakharova, E, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural roles of the Cys14-Cys38 disulfide of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 382, 2008
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
6MP3
 
 | |
2GC7
 
 | | Substrate reduced, copper free complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans. | | Descriptor: | Amicyanin, Cytochrome c-L, HEME C, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structral comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
4HX0
 
 | | Crystal structure of a putative nucleotidyltransferase (TM1012) from Thermotoga maritima at 1.87 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-BUTANEDIOL, Putative nucleotidyltransferase TM1012, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Lipkin, A.V, Popov, V.O, Kovalchuk, M.V, Shumilin, I.A, Minor, W, Shabalin, I.G, Golubev, A.M. | | Deposit date: | 2012-11-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of putative nucleotidyltransferase with two MPD molecules in the active site.
To be Published
|
|
1IVB
 
 | |
6MOZ
 
 | | Structure of acid-beta-glucosidase in complex with an aromatic pyrrolidine iminosugar inhibitor | | Descriptor: | (2R,3S,4R)-2-{[4-(3,5-dichlorophenyl)-1H-1,2,3-triazol-1-yl]methyl}pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Patterson-Orazem, A.C, Lieberman, R.L. | | Deposit date: | 2018-10-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring substituent diversity on pyrrolidine-aryltriazole iminosugars: Structural basis of beta-glucocerebrosidase inhibition.
Bioorg.Chem., 86, 2019
|
|
4CCY
 
 | | Crystal structure of carboxylesterase CesB (YbfK) from Bacillus subtilis | | Descriptor: | CARBOXYLESTERASE YBFK, SODIUM ION | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
3IHV
 
 | |
4HUS
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with virginiamycin M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
6N6K
 
 | | Human REXO2 bound to pAG | | Descriptor: | MALONATE ION, RNA (5'-R(P*AP*G)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, ... | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
