2AXQ
 
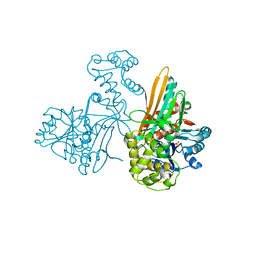 | |
4YV7
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL | | Descriptor: | GLYCEROL, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL
To be published
|
|
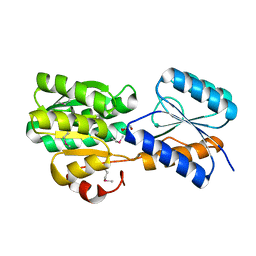
4R0N
 
 | | Hexagonal form of phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis | | Descriptor: | Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Timofeev, V.I, Smirnova, E.A, Chupova, L.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2014-08-01 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hexagonal form of phosphopantetheine adenylyltransferase from Mycobacterium tuberculosis
TO BE PUBLISHED
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
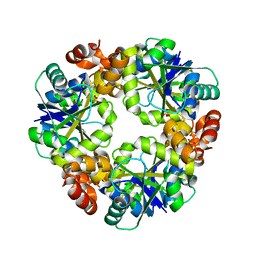
1R66
 
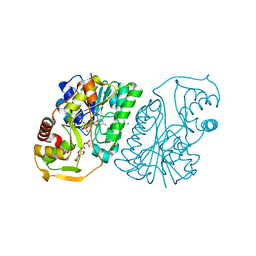 | | Crystal Structure of DesIV (dTDP-glucose 4,6-dehydratase) from Streptomyces venezuelae with NAD and TYD bound | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TDP-glucose-4,6-dehydratase, ... | | Authors: | Allard, S.T.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High Resolution X-ray Structure of dTDP-Glucose 4,6-Dehydratase from Streptomyces venezuelae
J.Biol.Chem., 279, 2004
|
|
6DNP
 
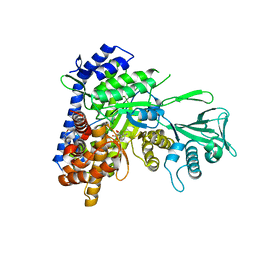 | | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 2-F-3-Methyl-6-F-phenyldiketoacid | | Descriptor: | (2Z)-4-(2,6-difluoro-3-methylphenyl)-2-hydroxy-4-oxobut-2-enoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Krieger, I.V, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC), Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Anion-pi Interactions in Computer-Aided Drug Design: Modeling the Inhibition of Malate Synthase by Phenyl-Diketo Acids.
J Chem Inf Model, 58, 2018
|
|
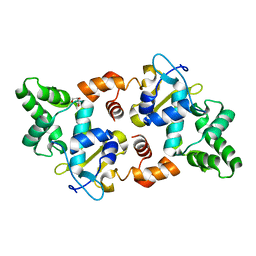
5AF3
 
 | |
7XOH
 
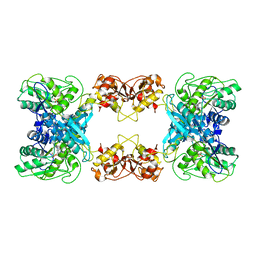 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine. | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5OU1
 
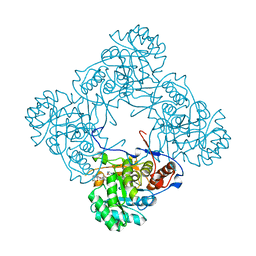 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 (7759844) | | Descriptor: | (2~{S})-~{N}-(4-iodophenyl)-2-(4-methoxyphenoxy)propanamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Based Approach to Targeting Inosine-5'-monophosphate Dehydrogenase (IMPDH) from Mycobacterium tuberculosis.
J. Med. Chem., 61, 2018
|
|
5OU3
 
 | | M. thermoresistible IMPDH in complex with IMP and Compound 31 (AT080) | | Descriptor: | (2~{S})-~{N}-[5-(4-bromophenyl)-1~{H}-imidazol-2-yl]-2-[4-(1-methylimidazol-4-yl)phenoxy]propanamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Approach to Targeting Inosine-5'-monophosphate Dehydrogenase (IMPDH) from Mycobacterium tuberculosis.
J. Med. Chem., 61, 2018
|
|
6KGS
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGW
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5HM3
 
 | | 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Warwrzak, Z, Kuhn, M.L, Shuvalova, L, Flores, K.J, Wilson, D.J, Grimes, K.D, Aldrich, C.C, Anderson, W.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-03 | | Last modified: | 2016-09-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Acs Infect Dis., 2, 2016
|
|
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | Descriptor: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6U7A
 
 | | Rv3722c in complex with kynurenine | | Descriptor: | (2S)-4-(2-aminophenyl)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-hydroxyquinoline-2-carboxylic acid, ... | | Authors: | Mandyoli, L, Sacchettini, J. | | Deposit date: | 2019-09-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis.
Nat Commun, 11, 2020
|
|
5OU2
 
 | | M. thermoresistible IMPDH in complex with IMP and Compound 2 (NMR744) | | Descriptor: | 4-(4-bromophenyl)-1H-imidazole, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fragment-Based Approach to Targeting Inosine-5'-monophosphate Dehydrogenase (IMPDH) from Mycobacterium tuberculosis.
J. Med. Chem., 61, 2018
|
|
6ULD
 
 | |
6U78
 
 | | Rv3722c in complex with glutamic acid | | Descriptor: | Aminotransferase, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Mandyoli, L, Sacchettini, J.C. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate aminotransferase Rv3722c governs aspartate-dependent nitrogen metabolism in Mycobacterium tuberculosis.
Nat Commun, 11, 2020
|
|
6URF
 
 | | Malic enzyme from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, NAD-dependent malic enzyme | | Authors: | Cuthbert, B.J, Burley, K.H, Goulding, C.W, Mathews, E.I, Beste, D.J. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Molecular Dynamics of Mycobacterium tuberculosis Malic Enzyme, a Potential Anti-TB Drug Target.
Acs Infect Dis., 7, 2021
|
|
5VO7
 
 | |
8IVP
 
 | | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase, D-fructose | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii
To Be Published
|
|
6KGV
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5IS2
 
 | |
7VJT
 
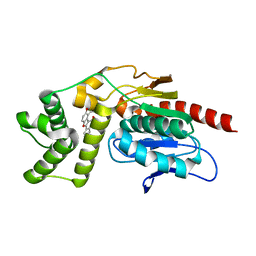 | | Crystal Structure of Mtb Pks13-TE in complex with inhibitor coumestan derivative 8 | | Descriptor: | 3,8-bis(oxidanyl)-7-(piperidin-1-ylmethyl)-[1]benzofuro[3,2-c]chromen-6-one, Polyketide synthase Pks13 (Termination polyketide synthase) | | Authors: | Zhang, W, Wang, S.S, Yu, L.F. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of Coumestan Derivatives as Polyketide Synthase 13-Thioesterase(Pks13-TE) Inhibitors with Improved hERG Profiles for Mycobacterium tuberculosis Treatment.
J.Med.Chem., 65, 2022
|
|
