2G31
 
 | | Human Nogo-A functional domain: nogo60 | | Descriptor: | Reticulon-4 | | Authors: | Li, M.F, Liu, J.X, Song, J.X. | | Deposit date: | 2006-02-17 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nogo goes in the pure water: solution structure of Nogo-60 and design of the structured and buffer-soluble Nogo-54 for enhancing CNS regeneration
Protein Sci., 15, 2006
|
|
2G32
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2G33
 
 | |
2G34
 
 | |
2G35
 
 | | NMR structure of talin-PTB in complex with PIPKI | | Descriptor: | Talin-1, peptide | | Authors: | Kong, X, Wang, X, Misra, S, Qin, J. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Phosphorylation-regulated Focal Adhesion Targeting of Type Igamma Phosphatidylinositol Phosphate Kinase (PIPKIgamma) by Talin.
J.Mol.Biol., 359, 2006
|
|
2G36
 
 | |
2G37
 
 | | Structure of Thermus thermophilus L-proline dehydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, proline dehydrogenase/delta-1-pyrroline-5-carboxylate dehydrogenase | | Authors: | Tanner, J.J, White, T.A. | | Deposit date: | 2006-02-17 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Kinetics of Monofunctional Proline Dehydrogenase from Thermus thermophilus.
J.Biol.Chem., 282, 2007
|
|
2G38
 
 | | A PE/PPE Protein Complex from Mycobacterium tuberculosis | | Descriptor: | MANGANESE (II) ION, PE FAMILY PROTEIN, PPE FAMILY PROTEIN | | Authors: | Strong, M, Sawaya, M.R, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G39
 
 | | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acetyl-CoA hydrolase | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa
To be Published
|
|
2G3A
 
 | | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | acetyltransferase | | Authors: | Cymborowski, M, Xu, X, Chruszcz, M, Zheng, H, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2G3B
 
 | | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | GLYCEROL, putative TetR-family transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Wang, S, Koclega, K.D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
2G3D
 
 | |
2G3F
 
 | | Crystal Structure of imidazolonepropionase complexed with imidazole-4-acetic acid sodium salt, a substrate homologue | | Descriptor: | 2H-IMIDAZOL-4-YLACETIC ACID, Imidazolonepropionase, ZINC ION | | Authors: | Yu, Y, Liang, Y.H, Su, X.D. | | Deposit date: | 2006-02-19 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A catalytic mechanism revealed by the crystal structures of the imidazolonepropionase from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
2G3H
 
 | | Cyanide Binding and Heme Cavity Conformational Transitions in Drosophila melanogaster Hexa-coordinate Hemoglobin | | Descriptor: | CHLORIDE ION, CYANIDE ION, MAGNESIUM ION, ... | | Authors: | de Sanctis, D, Ascenzi, P, Bocedi, A, Dewilde, S, Burmester, T, Hankeln, T, Moens, L, Bolognesi, M. | | Deposit date: | 2006-02-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyanide binding and heme cavity conformational transitions in Drosophila melanogaster hexacoordinate hemoglobin.
Biochemistry, 45, 2006
|
|
2G3I
 
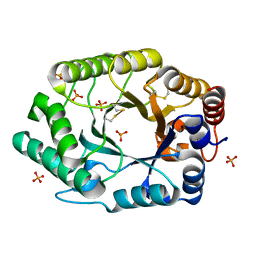 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
2G3J
 
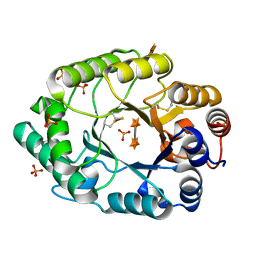 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase, alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
2G3K
 
 | | Crystal structure of the C-terminal domain of Vps28 | | Descriptor: | Vacuolar protein sorting-associated protein VPS28 | | Authors: | Pineda-Molina, E, Belrhali, H, Piefer, A.J, Akula, I, Bates, P, Weissenhorn, W. | | Deposit date: | 2006-02-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment.
Traffic, 7, 2006
|
|
2G3M
 
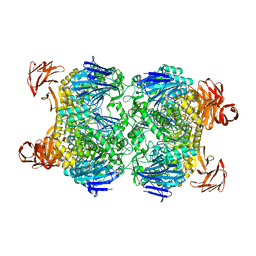 | | Crystal structure of the Sulfolobus solfataricus alpha-glucosidase MalA | | Descriptor: | Alpha-glucosidase | | Authors: | Ernst, H.A, Lo Leggio, L, Willemoes, M, Leonard, G, Blum, P, Larsen, S. | | Deposit date: | 2006-02-20 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Sulfolobus solfataricus alpha-Glucosidase: Implications for Domain Conservation and Substrate Recognition in GH31.
J.Mol.Biol., 358, 2006
|
|
2G3N
 
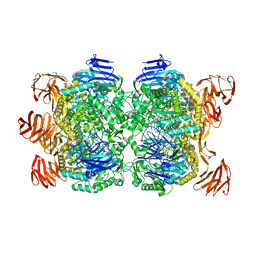 | | Crystal structure of the Sulfolobus solfataricus alpha-glucosidase MalA in complex with beta-octyl-glucopyranoside | | Descriptor: | Alpha-glucosidase, octyl beta-D-glucopyranoside | | Authors: | Ernst, H.A, Lo Leggio, L, Willemoes, M, Leonard, G, Blum, P, Larsen, S. | | Deposit date: | 2006-02-20 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Sulfolobus solfataricus alpha-Glucosidase: Implications for Domain Conservation and Substrate Recognition in GH31.
J.Mol.Biol., 358, 2006
|
|
2G3O
 
 | | The 2.1A crystal structure of copGFP | | Descriptor: | green fluorescent protein 2 | | Authors: | Wilmann, P.G. | | Deposit date: | 2006-02-20 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily
J.Mol.Biol., 359, 2006
|
|
2G3P
 
 | |
2G3Q
 
 | |
2G3R
 
 | | Crystal Structure of 53BP1 tandem tudor domains at 1.2 A resolution | | Descriptor: | SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
2G3S
 
 | | RNA structure containing GU base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3', MAGNESIUM ION | | Authors: | Jang, S.B, Hung, L.W, Jeong, M.S, Holbrook, E.L, Chen, X, Turner, D.H, Holbrook, S.R. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure at 1.5 angstroms resolution of an RNA octamer duplex containing tandem G.U basepairs
Biophys.J., 90, 2006
|
|
2G3T
 
 | |
