6S7P
 
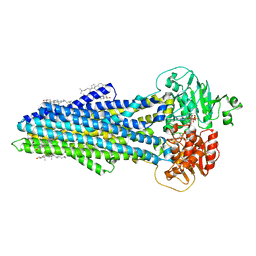 | | Nucleotide bound ABCB4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Olsen, J.A, Alam, A, Kowal, J, Stieger, B, Locher, K.P. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human lipid exporter ABCB4 in a lipid environment.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5WP9
 
 | | Structural Basis of Mitochondrial Receptor Binding and Constriction by Dynamin-Related Protein 1 | | Descriptor: | Dynamin-1-like protein, MAGNESIUM ION, Mitochondrial dynamics protein MID49, ... | | Authors: | Kalia, R, Wang, R.Y.R, Yusuf, A, Thomas, P.V, Agard, D.A, Shaw, J.M, Frost, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of mitochondrial receptor binding and constriction by DRP1.
Nature, 558, 2018
|
|
6K4J
 
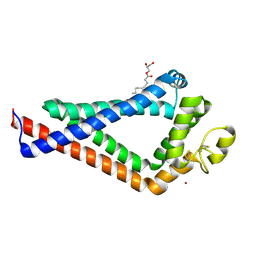 | | Crystal Structure of the the CD9 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | Authors: | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | Deposit date: | 2019-05-24 | | Release date: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
7A0V
 
 | | Crystal structure of the 5-phosphatase domain of Synaptojanin1 in complex with a nanobody | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nanobody 13015, ... | | Authors: | Paesmans, J, Galicia, C, Martin, E, Versees, W. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations.
Elife, 9, 2020
|
|
7A17
 
 | | Crystal structure of the 5-phosphatase domain of Synaptojanin1 bound to its substrate diC8-PI(3,4,5)P3 in complex with a nanobody | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, GLYCEROL, Isoform 2 of Synaptojanin-1, ... | | Authors: | Paesmans, J, Galicia, C, Martin, E, Versees, W. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations.
Elife, 9, 2020
|
|
5K9K
 
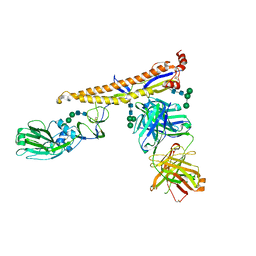 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 in complex with Hemagglutinin Hong Kong 1968. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 56.a.09 Heavy chain, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-31 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5XLA
 
 | |
5XL8
 
 | |
5XL9
 
 | |
6TKO
 
 | |
7AZP
 
 | |
6KZ8
 
 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6KZ9
 
 | | Crystal structure of plant Phospholipase D alpha | | Descriptor: | CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6UHC
 
 | | CryoEM structure of human Arp2/3 complex with bound NPFs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Zimmet, A, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2019-09-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism.
Sci Adv, 6, 2020
|
|
5L4Q
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase 1 (AAK1) in Complex with LKB1 (AAK1 Dual Inhibitor) | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, ~{N}-[5-(4-cyanophenyl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]pyridine-3-carboxamide | | Authors: | Sorrell, F.J, Williams, E, Fox, N, Abdul Azeez, K.R, Gileadi, O, von Delft, F, Edwards, A.M, Bountra, C, Elkins, J.M, Knapp, S. | | Deposit date: | 2016-05-26 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Synthesis and Structure-Activity Relationships of 3,5-Disubstituted-pyrrolo[2,3- b]pyridines as Inhibitors of Adaptor-Associated Kinase 1 with Antiviral Activity.
J.Med.Chem., 2019
|
|
7B4M
 
 | |
7B4L
 
 | |
5LOZ
 
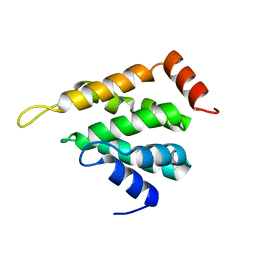 | | STRUCTURE OF YEAST ENT1 ENTH DOMAIN | | Descriptor: | Epsin-1 | | Authors: | Tanner, N, Prag, G. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A bacterial genetic selection system for ubiquitylation cascade discovery.
Nat.Methods, 13, 2016
|
|
7BY5
 
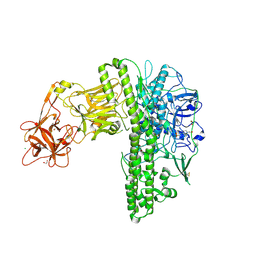 | | Tetanus neurotoxin mutant-(H233A/E234Q/H237A/Y375F) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Zhang, C.M, Imoto, Y, Fukuda, Y, Yamashita, E, Inoue, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X, 5, 2021
|
|
7BLO
 
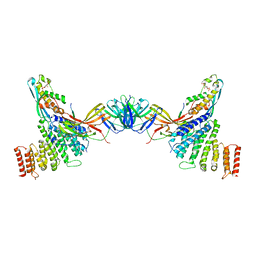 | | VPS26 dimer region of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, C-term (residues 493-54) of Wls (fitted sequence corresponds to hDMT1-II), Sorting nexin-3, ... | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
5LNP
 
 | |
6UYN
 
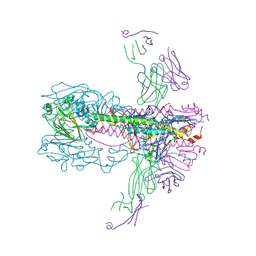 | |
6MSM
 
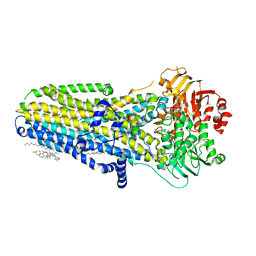 | | Phosphorylated, ATP-bound human cystic fibrosis transmembrane conductance regulator (CFTR) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Zhang, Z, Liu, F, Chen, J. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the ATP-bound, phosphorylated human CFTR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MC7
 
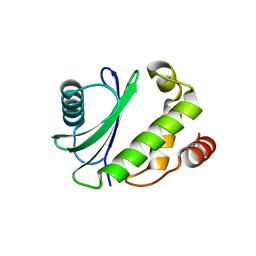 | |
6MRC
 
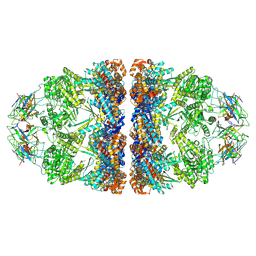 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
