9EPP
 
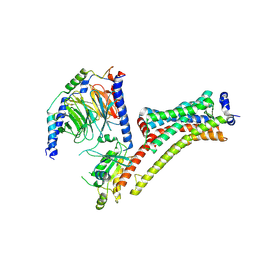 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Giq heterotrimer | | Descriptor: | 11,20-Ethanoretinal, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
9EP8
 
 | | Crystal structure of ROCK2 in complex with a 8-(azaindolyl)-benzoazepinone inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 8-(azaindolyl)-benzoazepinone, Rho-associated protein kinase 2 | | Authors: | Pala, D, Clark, D, Pioselli, B, Accetta, A, Rancati, F. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Design and synthesis of novel 8-(azaindolyl)-benzoazepinones as potent and selective ROCK inhibitors.
Rsc Med Chem, 2024
|
|
9B37
 
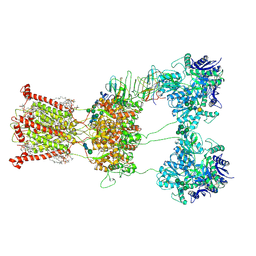 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B35
 
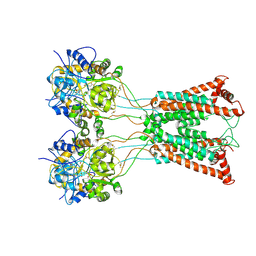 | | Ligand-binding and transmembrane domains of kainate receptor GluK2 in the open state, a complex with agonist glutamate and positive allosteric modulator BPAM344 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor ionotropic, kainate 2 | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B38
 
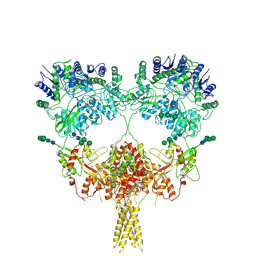 | | Kainate receptor GluK2 in complex with agonist glutamate with pseudo 4-fold symmetrical ligand-binding domain layer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B39
 
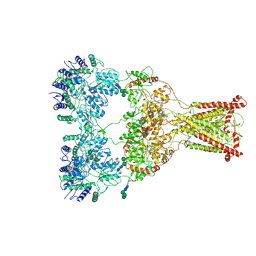 | | Kainate receptor GluK2 in complex with agonist glutamate with asymmetric ligand-binding domain layer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B36
 
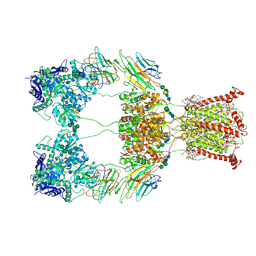 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9EOJ
 
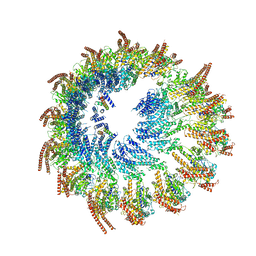 | | Vertebrate microtubule-capping gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component, Gamma-tubulin complex component 3 homolog, Gamma-tubulin complex component 6, ... | | Authors: | Vermeulen, B.J.A, Pfeffer, S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | gamma-TuRC asymmetry induces local protofilament mismatch at the RanGTP-stimulated microtubule minus end.
Embo J., 43, 2024
|
|
9B2H
 
 | | HIV-1 wild type protease with GRL-072-17A, a substituted tetrahydrofuran derivative based on Darunavir as P2 group | | Descriptor: | 2,5:6,9-dianhydro-1,3,7,8-tetradeoxy-4-O-({(2S,3R)-3-hydroxy-4-[(4-methoxybenzene-1-sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamoyl)-L-gluco-nonitol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2024-03-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Design of substituted tetrahydrofuran derivatives for HIV-1 protease inhibitors: synthesis, biological evaluation, and X-ray structural studies.
Org.Biomol.Chem., 22, 2024
|
|
9EO8
 
 | |
9EO2
 
 | |
9ENZ
 
 | |
9EO5
 
 | |
8YOY
 
 | | Structure of HKU1A RBD with TMPRSS2 | | Descriptor: | Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Shang, K, Zhu, H, Zhu, K. | | Deposit date: | 2024-03-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
9B27
 
 | | Dia1 at the Barbed End of F-Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-14 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9B1D
 
 | |
9B1E
 
 | |
9END
 
 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 3 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Coquille, S, Roche, J, Girard, E, Madern, D. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To be published
|
|
9B0K
 
 | | INF2 in the Middle of F-Actin (Down state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9AZI
 
 | |
9AZQ
 
 | | INF2 at the Barbed End of F-Actin with Incoming Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9B03
 
 | | INF2 in the Middle of F-Actin (Up state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9AZP
 
 | | INF2 at the Barbed End of F-Actin with Incoming Profilin-Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9AZL
 
 | | Cryo-EM structure of the ABC transporter PCAT1 bound with MgADP class1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
9AZK
 
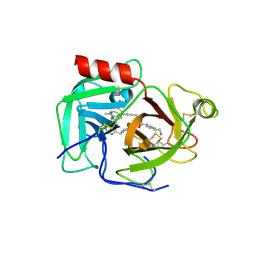 | | Macrocyclic inhibitors targeting the prime site of the fibrinolytic serine protease plasmin | | Descriptor: | (1r,4S)-4-(aminomethyl)-N-[(24S)-5-methyl-8,11,16,23-tetraoxo-7,10,15,22-tetraazatetracyclo[24.2.2.2~18,21~.1~2,6~]tritriaconta-1(28),2(33),3,5,18,20,26,29,31-nonaen-24-yl]cyclohexane-1-carboxamide, Plasminogen | | Authors: | Guojie, W. | | Deposit date: | 2024-03-11 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Macrocyclic inhibitors targeting the prime site of the fibrinolytic serine protease plasmin
To Be Published
|
|
