3GYJ
 
 | |
3GYI
 
 | |
3WWA
 
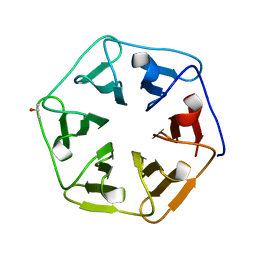 | | Crystal structure of the computationally designed Pizza7 protein after heat treatment | | Descriptor: | ISOPROPYL ALCOHOL, Pizza7H protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3EN3
 
 | | Crystal Structure of the GluR4 Ligand-Binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 4,Glutamate receptor | | Authors: | Gill, A, Madden, D.R. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists
Biochemistry, 47, 2008
|
|
4AW8
 
 | | X-ray structure of ZinT from Salmonella enterica in complex with zinc ion and PEG | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Metal-binding protein ZinT, SODIUM ION, ... | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
3WW9
 
 | | Crystal structure of the computationally designed Pizza6 protein | | Descriptor: | GLYCEROL, SULFATE ION, pizza6 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4AYH
 
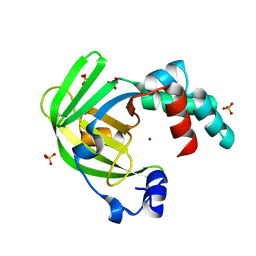 | | The X-ray structure of zinc bound ZinT | | Descriptor: | METAL-BINDING PROTEIN YODA, SULFATE ION, ZINC ION | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-06-21 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
4ARL
 
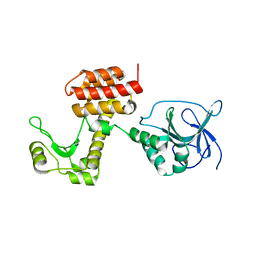 | | Structure of the inactive pesticin D207A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
3H3V
 
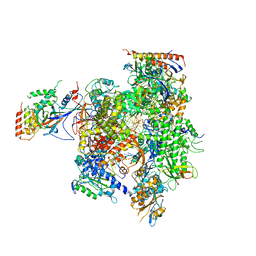 | | Yeast RNAP II containing poly(A)-signal sequence in the active site | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*GP*CP*TP*GP*CP*TP*TP*TP*AP*TP*TP*GP*CP*AP*TP*T)-3', 5'-D(*CP*AP*GP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*UP*CP*GP*CP*AP*AP*UP*AP*AP*A)-3', ... | | Authors: | Dengl, S, Cramer, P. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Torpedo Nuclease Rat1 Is Insufficient to Terminate RNA Polymerase II in Vitro
J.Biol.Chem., 284, 2009
|
|
3WW7
 
 | | Crystal structure of the computationally designed Pizza2 protein | | Descriptor: | GLYCEROL, Pizza2 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WG8
 
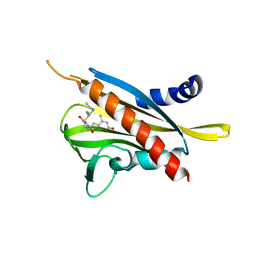 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist AS6 | | Descriptor: | (2Z,4E)-5-[(1S)-3-(hexylsulfanyl)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Okamoto, M, Muto, T, Endo, A, Nambara, E, Hirai, N, Ohnishi, T, Cutler, S.R, Todoroki, Y, Yajima, S. | | Deposit date: | 2013-07-31 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions
Nat.Chem.Biol., 10, 2014
|
|
4AUI
 
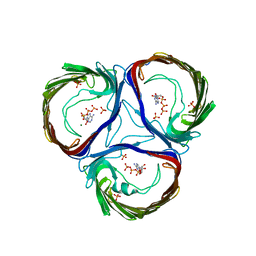 | | STRUCTURE AND FUNCTION OF THE PORB PORIN FROM DISSEMINATING N. GONORRHOEAE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Zeth, K, Kozjak-Pavlovic, V, Faulstich, M, Hurwitz, R, Kepp, O, Rudel, T. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Function of the Porb Porin from Disseminating Neisseria Gonorrhoeae.
Biochem.J., 449, 2013
|
|
4ARM
 
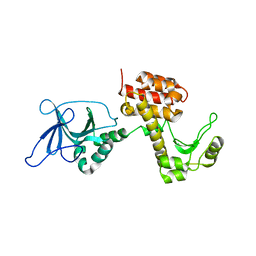 | | Structure of the inactive pesticin T201A mutant | | Descriptor: | PESTICIN | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
4GDE
 
 | |
4HOP
 
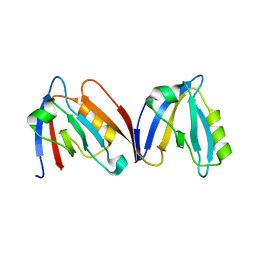 | | Crystal structure of the computationally designed NNOS-Syntrophin complex | | Descriptor: | Alpha-1-syntrophin, Nitric oxide synthase, brain | | Authors: | Harwood, I.M, Melero, C, Ollikainen, N, Kortemme, T. | | Deposit date: | 2012-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Quantification of the transferability of a designed protein specificity switch reveals extensive epistasis in molecular recognition.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4HPV
 
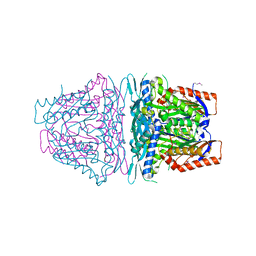 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-14 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4HIY
 
 | |
4HUD
 
 | | Structure of the bacteriophage T4 tail terminator protein, gp15. | | Descriptor: | Tail connector protein Gp15 | | Authors: | Fokine, A, Zhang, Z, Kanamaru, S, Bowman, V.D, Aksyuk, A, Arisaka, F, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2012-11-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | The molecular architecture of the bacteriophage t4 neck.
J.Mol.Biol., 425, 2013
|
|
4HI5
 
 | |
4I2Y
 
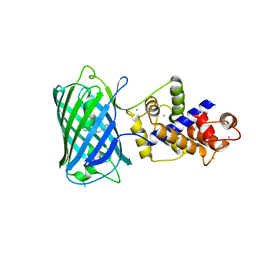 | |
4HBP
 
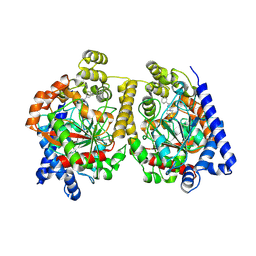 | | Crystal Structure of FAAH in complex with inhibitor | | Descriptor: | 4-(3-phenyl-1,2,4-thiadiazol-5-yl)-N-(pyridin-3-yl)piperazine-1-carboxamide, Fatty-acid amide hydrolase 1 | | Authors: | Behnke, C, Skene, R.J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of a series of piperazine ureas as fatty acid amide hydrolase (FAAH) inhibitors.
Bioorg.Med.Chem., 21, 2013
|
|
4FRF
 
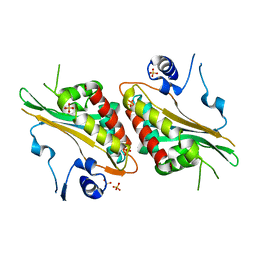 | | Structural Studies and Protein Engineering of Inositol Phosphate Multikinase | | Descriptor: | Inositol polyphosphate multikinase alpha, SULFATE ION | | Authors: | Endo-Streeter, S.T, Tsui, M, Odom, A.R, York, J.D. | | Deposit date: | 2012-06-26 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies and protein engineering of inositol phosphate multikinase.
J.Biol.Chem., 287, 2012
|
|
4FQN
 
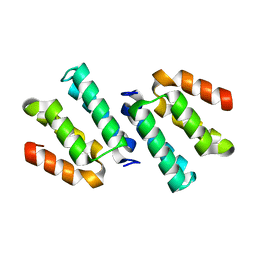 | | Crystal structure of the CCM2 C-terminal Harmonin Homology Domain (HHD) | | Descriptor: | Malcavernin | | Authors: | Fisher, O.S, Zhang, R, Li, X, Murphy, J.W, Boggon, T.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus.
Febs Lett., 587, 2013
|
|
4FZR
 
 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4HEI
 
 | | 2A X-RAY STRUCTURE OF HPF from VIBRIO CHOLERAE | | Descriptor: | COBALT (III) ION, RIBOSOME HIBERNATION PROTEIN YHBH, THIOCYANATE ION, ... | | Authors: | De Bari, H, Berry, E.A. | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Vibrio cholerae ribosome hibernation promoting factor.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
