5VXT
 
 | |
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
6ZPU
 
 | | Crystal structure of Angiotensin-1 converting enzyme C-domain with inserted symmetry molecule C-terminus. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Angiotensin-converting enzyme open for business: structural insights into the subdomain dynamics.
Febs J., 288, 2021
|
|
6ORV
 
 | | Non-peptide agonist (TT-OAD2) bound to the Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Danev, R. | | Deposit date: | 2019-05-01 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation of the GLP-1 receptor by a non-peptidic agonist.
Nature, 577, 2020
|
|
6OVL
 
 | |
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6OPN
 
 | | CD4- and 17-bound HIV-1 Env B41 SOSIP in complex with small molecule GO35 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
5VPF
 
 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | Authors: | Yin, Z, Rudenko, G, Machius, M. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VKC
 
 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
8YU6
 
 | | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Varfolomeeva, L.A, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of thiocyanate dehydrogenase mutant with the H447Q substitution from Pelomicrobium methylotrophicum (pmTcDH H447Q), activated by crystal soaking with 1mM CuCl2 and 1 mM sodium ascorbate
To Be Published
|
|
6OZ4
 
 | |
3JS2
 
 | | Crystal structure of minimal kinase domain of fibroblast growth factor receptor 1 in complex with 5-(2-thienyl)nicotinic acid | | Descriptor: | 5-(2-thienyl)nicotinic acid, Basic fibroblast growth factor receptor 1, PHOSPHATE ION | | Authors: | Bae, J.H, Ravindranathan, K.P, Mandiyan, V, Ekkati, A.R, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2009-09-09 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening
J.Med.Chem., 53, 2010
|
|
3KAO
 
 | | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SULFATE ION, Tagatose 1,6-diphosphate aldolase, ... | | Authors: | Chang, C, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus
To be Published
|
|
3CPX
 
 | |
3SNX
 
 | |
3SI9
 
 | | Crystal structure of Dihydrodipicolinate Synthase from Bartonella Henselae | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate synthase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Abendroth, J, Sankaran, B. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cloning, expression, purification, crystallization and X-ray diffraction analysis of dihydrodipicolinate synthase from the human pathogenic bacterium Bartonella henselae strain Houston-1 at 2.1 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7NXE
 
 | |
3SLI
 
 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH 2,7-ANHYDRO-NEU5AC PREPARED BY SOAKING WITH 3'-SIALYLLACTOSE | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Li, Y.T, Luo, M. | | Deposit date: | 1998-10-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A structures of leech intramolecular trans-sialidase complexes: evidence of its enzymatic mechanism.
J.Mol.Biol., 285, 1999
|
|
6OLP
 
 | | Full length HIV-1 Env AMC011 in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rantalainen, K, Cottrell, C.A. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics.
Plos Pathog., 15, 2019
|
|
3SWA
 
 | | E. Cloacae MurA R120A complex with UNAG and covalent adduct of PEP with CYS115 | | Descriptor: | 1,2-ETHANEDIOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Han, H, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
3K11
 
 | |
3CU9
 
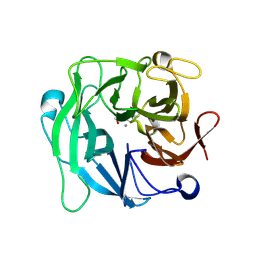 | | High resolution crystal structure of 1,5-alpha-L-arabinanase from Geobacillus Stearothermophilus | | Descriptor: | CALCIUM ION, GLYCEROL, Intracellular arabinanase | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-04-16 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
5VYC
 
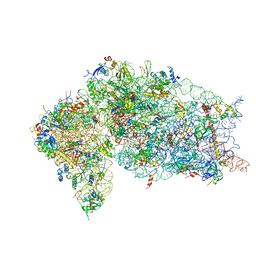 | | Crystal structure of the human 40S ribosomal subunit in complex with DENR-MCT-1. | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Lomakin, I.B, Stolboushkina, E.A, Vaidya, A.T, Garber, M.B, Dmitriev, S.E, Steitz, T.A. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structure of the Human Ribosome in Complex with DENR-MCT-1.
Cell Rep, 20, 2017
|
|
3T2T
 
 | |
3D4E
 
 | |
