2I3I
 
 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | (3R,6R,9AR)-2,2-DIMETHYL-6-[(N-METHYL-L-ALANYL)AMINO]-N-(3-METHYL-1-PHENYL-1H-PYRAZOL-5-YL)-5-OXO-2,3,5,6,9,9A-HEXAHYDRO[1,3]THIAZOLO[3,2-A]AZEPINE-3-CARBOXAMIDE, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Fairbrother, W.J, Franklin, M.C. | | Deposit date: | 2006-08-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs.
Acs Chem.Biol., 1, 2006
|
|
4WA8
 
 | |
4W87
 
 | | Crystal structure of XEG5A, a GH5 xyloglucan-specific endo-beta-1,4-glucanase from metagenomic library, in complex with a xyloglucan oligosaccharide | | Descriptor: | MAGNESIUM ION, Xyloglucan-specific endo-beta-1,4-glucanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4UVY
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methoxy-1,2- dihydroisoquinolin-1-one | | Descriptor: | 3-(4-chlorophenyl)-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
1EWE
 
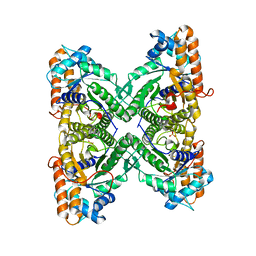 | | Fructose 1,6-Bisphosphate Aldolase from Rabbit Muscle | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE, SULFATE ION | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2000-04-25 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in
D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
4UDJ
 
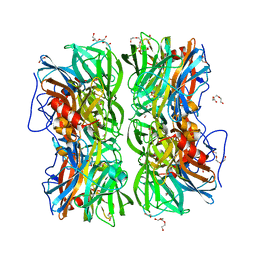 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.60 Angstrom in complex with beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UDK
 
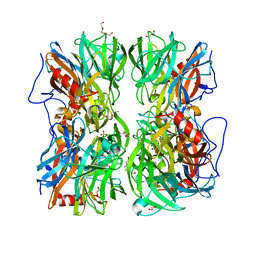 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.76 Angstrom from unknown human gut bacteria (Uhgb_MP) in complex with N-acetyl-D-glucosamine, beta-D-mannopyranose and inorganic phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1EJB
 
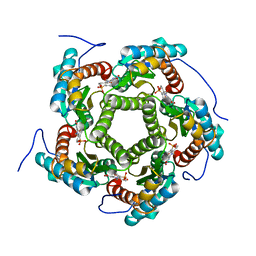 | | LUMAZINE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4-DIHYDROXYPYRIMIDIN-5-YL)-1-PENTYL-PHOSPHONIC ACID, LUMAZINE SYNTHASE | | Authors: | Meining, W, Mortl, S, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue.
J.Mol.Biol., 299, 2000
|
|
1ENT
 
 | |
1EWS
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE RABBIT KIDNEY DEFENSIN, RK-1 | | Descriptor: | RK-1 DEFENSIN | | Authors: | McManus, A.M, Dawson, N.F, Wade, J.D, Craik, D.J. | | Deposit date: | 2000-04-26 | | Release date: | 2001-05-02 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of RK-1: a novel alpha-defensin peptide.
Biochemistry, 39, 2000
|
|
1EYK
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH AMP, ZINC, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
1EC2
 
 | | HIV-1 protease in complex with the inhibitor BEA428 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DI-4-PYRIDIN-3-YL-BENZYL]-GLUCARYL]-DI-[VALYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EBW
 
 | | HIV-1 protease in complex with the inhibitor BEA322 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-[DIBENZYL]-GLUCARYL]-DI-[ISOLEUCYL-AMIDO-METHANE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EH5
 
 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 COMPLEXED WITH PALMITATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7AAZ
 
 | | Crystal structure of MerTK in complex with a type 1.5 aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-~{N}-[(1~{S},2~{S})-2-[[4-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]methoxy]cyclopentyl]-5-(1-methylpyrazol-4-yl)pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
7AAA
 
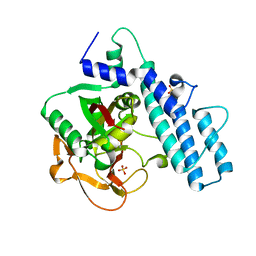 | | Crystal structure of the catalytic domain of human PARP1 (apo) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Underwood, E, Rawlins, P.B, Johannes, J.W, Easton, L.E, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
6ZMK
 
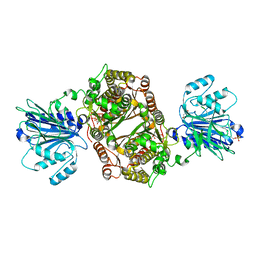 | | Crystal structure of human GFAT-1 L405R | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Mayr, F, Miethe, S, Atanassov, I, Baumann, U, Denzel, M.S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
6ZMJ
 
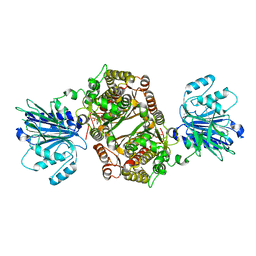 | | Crystal structure of human GFAT-1 R203H | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Mayr, F, Miethe, S, Atanassov, I, Baumann, U, Denzel, M.S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
5VT1
 
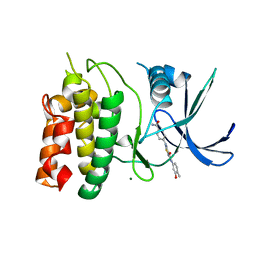 | | Crystal Structure of the Human CAMKK2B bound to a thiadiazinone benzamide inhibitor | | Descriptor: | 4-({5-[(3-hydroxy-4-methylphenyl)amino]-4-oxo-4H-1,2,6-thiadiazin-3-yl}amino)benzamide, Calcium/calmodulin-dependent protein kinase kinase 2, MAGNESIUM ION | | Authors: | Counago, R.M, Asquith, C.R.M, Arruda, P, Edwards, A.M, Gileadi, O, Kalogirou, A.S, Koutentis, P.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-15 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,2,6-Thiadiazinones as Novel Narrow Spectrum Calcium/Calmodulin-Dependent Protein Kinase Kinase 2 (CaMKK2) Inhibitors.
Molecules, 23, 2018
|
|
6ZQ1
 
 | | Structure of AraDNJ-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5VYS
 
 | | Crystal structure of the WbkC N-formyltransferase (C47S variant) from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
6ZPQ
 
 | |
6ZPT
 
 | |
5VVF
 
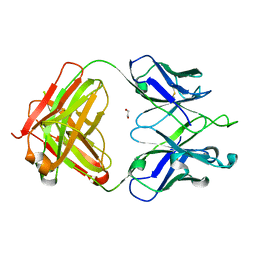 | | Crystal Structure of 354BG1 Fab | | Descriptor: | 1,2-ETHANEDIOL, 354BG1 Heavy Chain, 354BG1 Light Chain | | Authors: | Scharf, L, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-05-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies.
Elife, 6, 2017
|
|
6ZYM
 
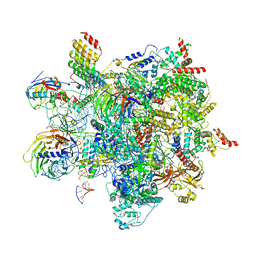 | | Human C Complex Spliceosome - High-resolution CORE | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Corepressor interacting with RBPJ 1, ... | | Authors: | Bertram, K, Kastner, B. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into the Roles of Metazoan-Specific Splicing Factors in the Human Step 1 Spliceosome.
Mol.Cell, 80, 2020
|
|
