6YF8
 
 | | DYRK1A with PST001 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}-[5-(5-methoxy-1,3-benzothiazol-2-yl)pyridin-3-yl]ethanamide | | Authors: | Rothweiler, U. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | DYRK1A with PST001
To Be Published
|
|
8T0S
 
 | | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Wang, C, Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position
To be published
|
|
6YGU
 
 | | Crystal structure of the minimal Mtr4-Red1 complex (single chain) from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ATP dependent RNA helicase (Dob1)-like protein, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
8TCK
 
 | |
8Z56
 
 | |
6OCD
 
 | | Ricin A chain bound to VHH antibody V6D4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2019-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Intracellular Neutralization of Ricin Toxin by Single-domain Antibodies Targeting the Active Site.
J.Mol.Biol., 432, 2020
|
|
5NSA
 
 | | Beta domain of human transcobalamin bound to Co-beta-[2-(2,4-difluorophenyl)ethinyl]cobalamin | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, CALCIUM ION, COBALAMIN, ... | | Authors: | Bloch, J.S, Ruetz, M, Krautler, B, Locher, K.P. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.273 Å) | | Cite: | Structure of the human transcobalamin beta domain in four distinct states.
PLoS ONE, 12, 2017
|
|
8RHO
 
 | | [2Fe:2S] ferredoxin FeSII from Azotobacter vinelandii, reduced form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Franke, P, Zhang, L, Einsle, O. | | Deposit date: | 2023-12-15 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational protection of molybdenum nitrogenase by Shethna protein II.
Nature, 637, 2025
|
|
8DKO
 
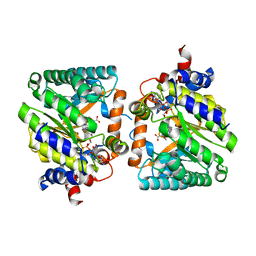 | |
6QNC
 
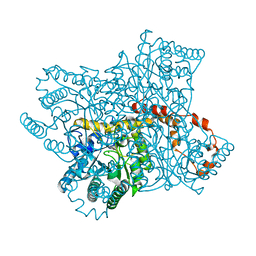 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 0.1 s timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6VRF
 
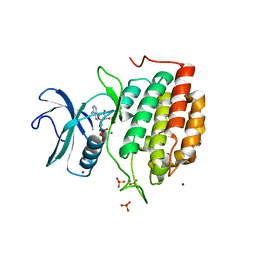 | | ADP bound TTBK2 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanisms of Regulation and Diverse Activities of Tau-Tubulin Kinase (TTBK) Isoforms.
Cell Mol Neurobiol, 41, 2021
|
|
8TZD
 
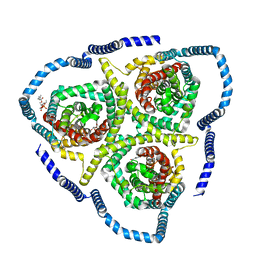 | |
8DLB
 
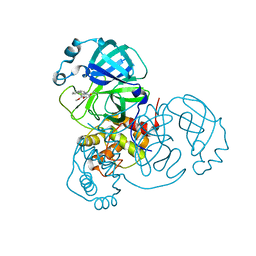 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | Descriptor: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
8DMD
 
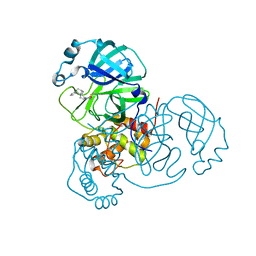 | |
7RTI
 
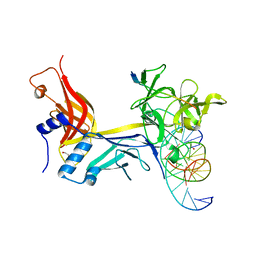 | | X-ray structure of RBPJ-L3MBTL3(dT62)-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Hall, D.P, Kovall, R.A, Yuan, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure, binding and function of a Notch transcription complex involving RBPJ and the epigenetic reader protein L3MBTL3.
Nucleic Acids Res., 50, 2022
|
|
8TZ7
 
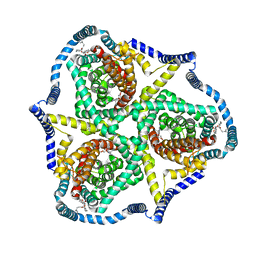 | |
8G4J
 
 | |
8TZ8
 
 | |
7BWQ
 
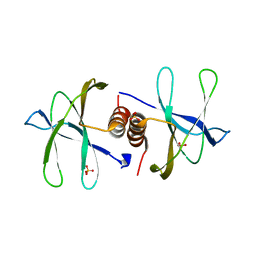 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
6OIP
 
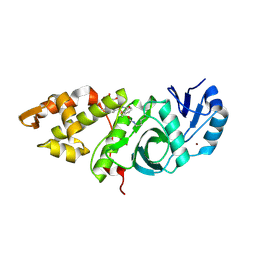 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 34 | | Descriptor: | 2-fluoro-3-methyl-N'-[(naphthalen-2-yl)sulfonyl]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
4XME
 
 | |
8ECG
 
 | | Complex of HMG1 with pitavastatin | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1, GLYCEROL, Pitavastatin | | Authors: | Haywood, J, Bond, C.S. | | Deposit date: | 2022-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.
Nat Commun, 13, 2022
|
|
8UB5
 
 | | The Apo NanH structure from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, Sialidase | | Authors: | Medley, B.J, Boraston, A.B. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
7S1N
 
 | |
4XMH
 
 | |
